Functional refolding of the penetration protein on a non-enveloped virus
- PMID: 33442061
- PMCID: PMC8297411
- DOI: 10.1038/s41586-020-03124-4
Functional refolding of the penetration protein on a non-enveloped virus
Abstract
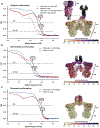
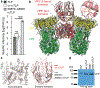
A non-enveloped virus requires a membrane lesion to deliver its genome into a target cell1. For rotaviruses, membrane perforation is a principal function of the viral outer-layer protein, VP42,3. Here we describe the use of electron cryomicroscopy to determine how VP4 performs this function and show that when activated by cleavage to VP8* and VP5*, VP4 can rearrange on the virion surface from an 'upright' to a 'reversed' conformation. The reversed structure projects a previously buried 'foot' domain outwards into the membrane of the host cell to which the virion has attached. Electron cryotomograms of virus particles entering cells are consistent with this picture. Using a disulfide mutant of VP4, we have also stabilized a probable intermediate in the transition between the two conformations. Our results define molecular mechanisms for the first steps of the penetration of rotaviruses into the membranes of target cells and suggest similarities with mechanisms postulated for other viruses.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures








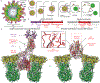
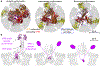

Similar articles
-
Atomic model of an infectious rotavirus particle.EMBO J. 2011 Jan 19;30(2):408-16. doi: 10.1038/emboj.2010.322. Epub 2010 Dec 14. EMBO J. 2011. PMID: 21157433 Free PMC article.
-
Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.J Virol. 2022 Aug 24;96(16):e0062722. doi: 10.1128/jvi.00627-22. Epub 2022 Aug 4. J Virol. 2022. PMID: 35924923 Free PMC article.
-
Visualization of Calcium Ion Loss from Rotavirus during Cell Entry.J Virol. 2018 Nov 27;92(24):e01327-18. doi: 10.1128/JVI.01327-18. Print 2018 Dec 15. J Virol. 2018. PMID: 30258012 Free PMC article.
-
Rotavirus Replication: Gaps of Knowledge on Virus Entry and Morphogenesis.Tohoku J Exp Med. 2019 Aug;248(4):285-296. doi: 10.1620/tjem.248.285. Tohoku J Exp Med. 2019. PMID: 31447474 Review.
-
Genetic diversity of G1P[8] rotavirus VP7 and VP8* antigens in Finland over a 20-year period: No evidence for selection pressure by universal mass vaccination with RotaTeq® vaccine.Infect Genet Evol. 2013 Oct;19:51-8. doi: 10.1016/j.meegid.2013.06.026. Epub 2013 Jul 4. Infect Genet Evol. 2013. PMID: 23831933 Review.
Cited by
-
mRNA-based VP8* nanoparticle vaccines against rotavirus are highly immunogenic in rodents.NPJ Vaccines. 2023 Dec 22;8(1):190. doi: 10.1038/s41541-023-00790-z. NPJ Vaccines. 2023. PMID: 38129390 Free PMC article.
-
Mechanisms of Cell Entry by dsRNA Viruses: Insights for Efficient Delivery of dsRNA and Tools for Improved RNAi-Based Pest Control.Front Physiol. 2021 Nov 11;12:749387. doi: 10.3389/fphys.2021.749387. eCollection 2021. Front Physiol. 2021. PMID: 34858204 Free PMC article. Review.
-
Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.Nat Commun. 2024 Mar 13;15(1):2284. doi: 10.1038/s41467-024-46624-x. Nat Commun. 2024. PMID: 38480794 Free PMC article.
-
VP4 Is a Determinant of Alpha-Defensin Modulation of Rotaviral Infection.J Virol. 2022 Apr 13;96(7):e0205321. doi: 10.1128/jvi.02053-21. Epub 2022 Mar 14. J Virol. 2022. Corrected and republished in: J Virol. 2023 Oct 31;97(10):e0096223. doi: 10.1128/jvi.00962-23. PMID: 35285683 Free PMC article. Corrected and republished.
-
The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.bioRxiv [Preprint]. 2023 Oct 16:2023.10.15.562449. doi: 10.1101/2023.10.15.562449. bioRxiv. 2023. Update in: PLoS Pathog. 2024 Apr 4;20(4):e1011750. doi: 10.1371/journal.ppat.1011750. PMID: 37905109 Free PMC article. Updated. Preprint.
References
-
- Harrison SC in Fields Virology (6th edition) (ed Knipe DM and Howley PM) 52–86 (Wolters Kluwer/Lippincott Williams and Wilkins, 2013).
-
- Estes MK & Greenberg H in Fields Virology (6th edition) (ed Knipe DM and Howley PM) 1347–1401 (Wolters Kluwer/Lippincott Williams and Wilkins, 2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

