Chaperone-mediated autophagy sustains haematopoietic stem-cell function
- PMID: 33442062
- PMCID: PMC8428053
- DOI: 10.1038/s41586-020-03129-z
Chaperone-mediated autophagy sustains haematopoietic stem-cell function
Abstract
The activation of mostly quiescent haematopoietic stem cells (HSCs) is a prerequisite for life-long production of blood cells1. This process requires major molecular adaptations to allow HSCs to meet the regulatory and metabolic requirements for cell division2-4. The mechanisms that govern cellular reprograming upon stem-cell activation, and the subsequent return of stem cells to quiescence, have not been fully characterized. Here we show that chaperone-mediated autophagy (CMA)5, a selective form of lysosomal protein degradation, is involved in sustaining HSC function in adult mice. CMA is required for protein quality control in stem cells and for the upregulation of fatty acid metabolism upon HSC activation. We find that CMA activity in HSCs decreases with age and show that genetic or pharmacological activation of CMA can restore the functionality of old mouse and human HSCs. Together, our findings provide mechanistic insights into a role for CMA in sustaining quality control, appropriate energetics and overall long-term HSC function. Our work suggests that CMA may be a promising therapeutic target for enhancing HSC function in conditions such as ageing or stem-cell transplantation.
Conflict of interest statement
Competing interest
AMC and EG are cofounders of Selphagy Therapeutics (Boston, MA, USA).
Figures



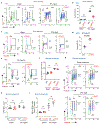



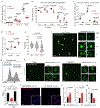
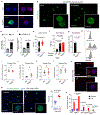

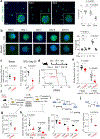


Comment in
-
Young haematopoietic stem cells are picky eaters.Cell Res. 2021 Apr;31(4):377-378. doi: 10.1038/s41422-021-00488-8. Cell Res. 2021. PMID: 33633351 Free PMC article. No abstract available.
References
-
- Jordan CT & Lemischka IR Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev 4, 220–232 (1990). - PubMed
-
- Cabezas-Wallscheid N et al.Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15, 507–522 (2014). - PubMed
-
- Arai F & Suda T Maintenance of quiescent hematopoietic stem cells in the osteoblastic niche. Ann N Y Acad Sci 1106, 41–53 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL146442/HL/NHLBI NIH HHS/United States
- R01 AG021904/AG/NIA NIH HHS/United States
- K01 DK105134/DK/NIDDK NIH HHS/United States
- T32 AG023475/AG/NIA NIH HHS/United States
- DK105134/NH/NIH HHS/United States
- R37 AG021904/AG/NIA NIH HHS/United States
- AG023475/NH/NIH HHS/United States
- P30 AG038072/AG/NIA NIH HHS/United States
- R21 HL150032/HL/NHLBI NIH HHS/United States
- R01 HL149714/HL/NHLBI NIH HHS/United States
- P30 CA013330/CA/NCI NIH HHS/United States
- P01 AG031782/AG/NIA NIH HHS/United States
- R01 CA230756/CA/NCI NIH HHS/United States
- R01 HL148151/HL/NHLBI NIH HHS/United States
- R01 DK098408/DK/NIDDK NIH HHS/United States
- CA330756/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

