Pivotal role of long non-coding ribonucleic acid-X-inactive specific transcript in regulating immune checkpoint programmed death ligand 1 through a shared pathway between miR-194-5p and miR-155-5p in hepatocellular carcinoma
- PMID: 33442449
- PMCID: PMC7772730
- DOI: 10.4254/wjh.v12.i12.1211
Pivotal role of long non-coding ribonucleic acid-X-inactive specific transcript in regulating immune checkpoint programmed death ligand 1 through a shared pathway between miR-194-5p and miR-155-5p in hepatocellular carcinoma
Abstract
Background: Anti-programmed death therapy has thrust immunotherapy into the spotlight. However, such therapy has a modest response in hepatocellular carcinoma (HCC). Epigenetic immunomodulation is a suggestive combinatorial therapy with immune checkpoint blockade. Non-coding ribonucleic acid (ncRNA) driven regulation is a major mechanism of epigenetic modulation. Given the wide range of ncRNAs that co-opt in programmed cell-death protein 1 (PD-1)/programmed death ligand 1 (PD-L1) regulation, and based on the literature, we hypothesized that miR-155-5p, miR-194-5p and long non-coding RNAs (lncRNAs) X-inactive specific transcript (XIST) and MALAT-1 are involved in a regulatory upstream pathway for PD-1/PD-L1. Recently, nutraceutical therapeutics in cancers have received increasing attention. Thus, it is interesting to study the impact of oleuropein on the respective study key players.
Aim: To explore potential upstream regulatory ncRNAs for the immune checkpoint PD-1/PD-L1.
Methods: Bioinformatics tools including microrna.org and lnCeDB software were adopted to detect targeting of miR-155-5p, miR-194-5p and lncRNAs XIST and MALAT-1 to PD-L1 mRNA, respectively. In addition, Diana tool was used to predict targeting of both aforementioned miRNAs to lncRNAs XIST and MALAT-1. HCC and normal tissue samples were collected for scanning of PD-L1, XIST and MALAT-1 expression. To study the interaction among miR-155-5p, miR-194-5p, lncRNAs XIST and MALAT-1, as well as PD-L1 mRNA, a series of transfections of the Huh-7 cell line was carried out.
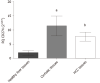
Results: Bioinformatics software predicted that miR-155-5p and miR-194-5p can target PD-L1, MALAT-1 and XIST. MALAT-1 and XIST were predicted to target PD-L1 mRNA. PD-L1 and XIST were significantly upregulated in 23 HCC biopsies compared to healthy controls; however, MALAT-1 was barely detected. MiR-194 induced expression elevated the expression of PD-L1, XIST and MALAT-1. However, overexpression of miR-155-5p induced the upregulation of PD-L1 and XIST, while it had a negative impact on MALAT-1 expression. Knockdown of XIST did have an impact on PD-L1 expression; however, following knockdown of the negative regulator of X-inactive specific transcript (TSIX), PD-L1 expression was elevated, and abolished MALAT-1 activity. Upon co-transfection of miR-194-5p with siMALAT-1, PD-L1 expression was elevated. Co-transfection of miR-194-5p with siXIST did not have an impact on PD-L1 expression. Upon co-transfection of miR-194 with siTSIX, PD-L1 expression was upregulated. Interestingly, the same PD-L1 expression pattern was observed following miR-155-5p co-transfections. Oleuropein treatment of Huh-7 cells reduced the expression profile of PD-L1, XIST, and miR-155-5p, upregulated the expression of miR-194-5p and had no significant impact on the MALAT-1 expression profile.
Conclusion: This study reported a novel finding revealing that opposing acting miRNAs in HCC, have the same impact on PD-1/PD-L1 immune checkpoint by sharing a common signaling pathway.
Keywords: Hepatocellular carcinoma; Immune checkpoint; MiR-155-5p; MiR-194-5p; Programmed cell-death protein 1/Programmed death ligand 1; X-inactive specific transcript.
©The Author(s) 2020. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors have no conflicts of interests.
Figures








References
-
- Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408–424. - PubMed
-
- Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

