This is a preprint.
Lipidomic Signatures Align with Inflammatory Patterns and Outcomes in Critical Illness
- PMID: 33442677
- PMCID: PMC7805459
- DOI: 10.21203/rs.3.rs-106579/v1
Lipidomic Signatures Align with Inflammatory Patterns and Outcomes in Critical Illness
Update in
-
Lipidomic signatures align with inflammatory patterns and outcomes in critical illness.Nat Commun. 2022 Nov 10;13(1):6789. doi: 10.1038/s41467-022-34420-4. Nat Commun. 2022. PMID: 36357394 Free PMC article.
Abstract
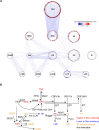
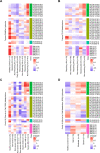
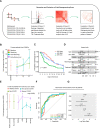
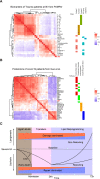
Alterations in lipid metabolism have the potential to be markers as well as drivers of the pathobiology of acute critical illness. Here, we took advantage of the temporal precision offered by trauma as a common cause of critical illness to identify the dynamic patterns in the circulating lipidome in critically ill humans. The major findings include an early loss of all classes of circulating lipids followed by a delayed and selective lipogenesis in patients destined to remain critically ill. Early in the clinical course, Fresh Frozen Plasma administration led to improved survival in association with preserved lipid levels that related to favorable changes in coagulation and inflammation biomarkers. Late over-representation of phosphatidylethanolamines with critical illness led to the validation of a Lipid Reprogramming Score that was prognostic not only in trauma but also severe COVID-19 patients. Our lipidomic findings provide a new paradigm for the lipid response underlying critical illness.
Conflict of interest statement
Declaration of Interests.
The authors declare no competing interests.
Figures



References
-
- Davidson G. H. et al. Long-term survival of adult trauma patients. JAMA 305, 1001–1007 (2011). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

