This is a preprint.
SARS-CoV-2 induces human plasmacytoid pre-dendritic cell diversification via UNC93B and IRAK4
- PMID: 33442685
- PMCID: PMC7805442
- DOI: 10.1101/2020.07.10.197343
SARS-CoV-2 induces human plasmacytoid pre-dendritic cell diversification via UNC93B and IRAK4
Update in
-
SARS-CoV-2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4.J Exp Med. 2021 Apr 5;218(4):e20201387. doi: 10.1084/jem.20201387. J Exp Med. 2021. PMID: 33533916 Free PMC article.
Abstract
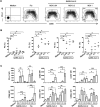
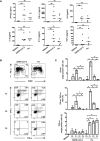
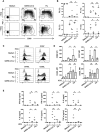
Several studies have analyzed antiviral immune pathways in late-stage severe COVID-19. However, the initial steps of SARS-CoV-2 antiviral immunity are poorly understood. Here, we have isolated primary SARS-CoV-2 viral strains, and studied their interaction with human plasmacytoid pre-dendritic cells (pDC), a key player in antiviral immunity. We show that pDC are not productively infected by SARS-CoV-2. However, they efficiently diversified into activated P1-, P2-, and P3-pDC effector subsets in response to viral stimulation. They expressed CD80, CD86, CCR7, and OX40 ligand at levels similar to influenza virus-induced activation. They rapidly produced high levels of interferon-α, interferon-λ1, IL-6, IP-10, and IL-8. All major aspects of SARS-CoV-2-induced pDC activation were inhibited by hydroxychloroquine. Mechanistically, SARS-CoV-2-induced pDC activation critically depended on IRAK4 and UNC93B1, as established using pDC from genetically deficient patients. Overall, our data indicate that human pDC are efficiently activated by SARS-CoV-2 particles and may thus contribute to type I IFN-dependent immunity against SARS-CoV-2 infection.
Conflict of interest statement
Competing interest
The authors declare no competing interests.
Figures


References
-
- Alculumbre S.G., Saint-André V., Di Domizio J., Vargas P., Sirven P., Bost P., Maurin M., Maiuri P., Wery M., Roman M.S., Savey L., Touzot M., Terrier B., Saadoun D., Conrad C., Gilliet M., Morillon A., and Soumelis V.. 2018. Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat. Immunol. 19:63–75. doi: 10.1038/s41590-017-0012-z - DOI - PubMed
-
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., Manry J., Shaw E., Haljasmägi L., Peterson P., Lorenzo L., Bizien L., Trouillet-Assant S., Dobbs K., de Jesus A.A., Belot A., Kallaste A., Catherinot E., Tandjaoui-Lambiotte Y., Le Pen J., Kerner G., Bigio B., Seeleuthner Y., Yang R., Bolze A., Spaan A.N., Delmonte O.M., Abers M.S., Aiuti A., Casari G., Lampasona V., Piemonti L., Ciceri F., Bilguvar K., Lifton R.P., Vasse M., Smadja D.M., Migaud M., Hadjadj J., Terrier B., Duffy D., Quintana-Murci L., van de Beek D., Roussel L., Vinh D.C., Tangye S.G., Haerynck F., Dalmau D., Martinez-Picado J., Brodin P., Nussenzweig M.C., Boisson-Dupuis S., Rodríguez-Gallego C., Vogt G., Mogensen T.H., Oler A.J., Gu J., Burbelo P.D., Cohen J.I., Biondi A., Bettini L.R., D’Angio M., Bonfanti P., Rossignol P., Mayaux J., Rieux-Laucat F., Husebye E.S., Fusco F., Ursini M.V., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Castagnoli R., Montagna D., Licari A., Marseglia G.L., Duval X., Ghosn J., HGID Lab, NIAID-USUHS Immune Response to COVID Group, COVID Clinicians, COVID-STORM Clinicians, Imagine COVID Group, French COVID Cohort Study Group, Milieu Intérieur Consortium, CoV-Contact Cohort, Amsterdam UMC Covid-19 Biobank, COVID Human Genetic Effort, Tsang J.S., Goldbach-Mansky R., Kisand K., Lionakis M.S., et al. 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 370. doi: 10.1126/science.abd4585 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
