This is a preprint.
Molecular dynamics simulations and functional studies reveal that hBD-2 binds SARS-CoV-2 spike RBD and blocks viral entry into ACE2 expressing cells
- PMID: 33442698
- PMCID: PMC7805467
- DOI: 10.1101/2021.01.07.425621
Molecular dynamics simulations and functional studies reveal that hBD-2 binds SARS-CoV-2 spike RBD and blocks viral entry into ACE2 expressing cells
Abstract
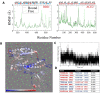
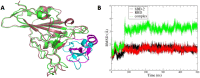
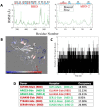
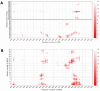
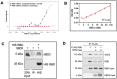
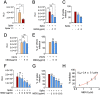
New approaches to complement vaccination are needed to combat the spread of SARS-CoV-2 and stop COVID-19 related deaths and long-term medical complications. Human beta defensin 2 (hBD-2) is a naturally occurring epithelial cell derived host defense peptide that has antiviral properties. Our comprehensive in-silico studies demonstrate that hBD-2 binds the site on the CoV-2-RBD that docks with the ACE2 receptor. Biophysical and biochemical assays confirm that hBD-2 indeed binds to the CoV-2-receptor binding domain (RBD) (KD ~ 300 nM), preventing it from binding to ACE2 expressing cells. Importantly, hBD-2 shows specificity by blocking CoV-2/spike pseudoviral infection, but not VSV-G mediated infection, of ACE2 expressing human cells with an IC50 of 2.4± 0.1 μM. These promising findings offer opportunities to develop hBD-2 and/or its derivatives and mimetics to safely and effectively use as novel agents to prevent SARS-CoV-2 infection.
Keywords: ACE2 receptor; COVID-19; Human beta defensin-2 (hBD-2); SARS-CoV-2; receptor binding domain (RBD).
Conflict of interest statement
DECLARATION OF INTERESTS: None to declare
Figures
Similar articles
-
HBD-2 binds SARS-CoV-2 RBD and blocks viral entry: Strategy to combat COVID-19.iScience. 2022 Mar 18;25(3):103856. doi: 10.1016/j.isci.2022.103856. Epub 2022 Feb 2. iScience. 2022. PMID: 35128350 Free PMC article.
-
Exploring the Interaction of RBD with Human β Defensin Type 2 Point Mutants: Insights from Molecular Dynamics Simulations.J Phys Chem B. 2025 Feb 20;129(7):1927-1933. doi: 10.1021/acs.jpcb.4c07004. Epub 2025 Feb 10. J Phys Chem B. 2025. PMID: 39929747
-
Molecular Dynamics Simulations on Spike Protein Mutants Binding with Human β Defensin Type 2.J Phys Chem B. 2024 Jan 18;128(2):415-428. doi: 10.1021/acs.jpcb.3c05460. Epub 2024 Jan 8. J Phys Chem B. 2024. PMID: 38189674
-
Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2-ACE2 Receptor Interactions.mBio. 2021 Mar 30;12(2):e03681-20. doi: 10.1128/mBio.03681-20. mBio. 2021. PMID: 33785634 Free PMC article.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
References
-
- Bensch K.W., Raida M., Mägert H.J., Schulz-Knappe P., and Forssmann W.G. (1995). hBD-1: a novel beta-defensin from human plasma. FEBS Lett 368, 331–335. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
