SP3-FAIMS Chemoproteomics for High-Coverage Profiling of the Human Cysteinome*
- PMID: 33442901
- PMCID: PMC8942723
- DOI: 10.1002/cbic.202000870
SP3-FAIMS Chemoproteomics for High-Coverage Profiling of the Human Cysteinome*
Abstract
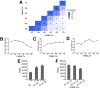
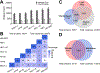
Chemoproteomics has enabled the rapid and proteome-wide discovery of functional, redox-sensitive, and ligandable cysteine residues. Despite widespread adoption and considerable advances in both sample-preparation workflows and MS instrumentation, chemoproteomics experiments still typically only identify a small fraction of all cysteines encoded by the human genome. Here, we develop an optimized sample-preparation workflow that combines enhanced peptide labeling with single-pot, solid-phase-enhanced sample-preparation (SP3) to improve the recovery of biotinylated peptides, even from small sample sizes. By combining this improved workflow with on-line high-field asymmetric waveform ion mobility spectrometry (FAIMS) separation of labeled peptides, we achieve unprecedented coverage of >14000 unique cysteines in a single-shot 70 min experiment. Showcasing the wide utility of the SP3-FAIMS chemoproteomic method, we find that it is also compatible with competitive small-molecule screening by isotopic tandem orthogonal proteolysis-activity-based protein profiling (isoTOP-ABPP). In aggregate, our analysis of 18 samples from seven cell lines identified 34225 unique cysteines using only ∼28 h of instrument time. The comprehensive spectral library and improved coverage provided by the SP3-FAIMS chemoproteomics method will provide the technical foundation for future studies aimed at deciphering the functions and druggability of the human cysteineome.
Keywords: LC-MS/MS; chemoproteomics; cysteine; high-field asymmetric waveform ion mobility spectrometry (FAIMS); single-pot, solid-phase-enhanced sample-preparation (SP3).
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures
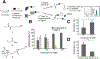


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

