Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence
- PMID: 33443036
- PMCID: PMC7877496
- DOI: 10.1126/sciimmunol.abf8891
Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence
Abstract
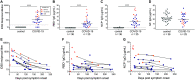
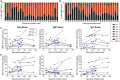
Lasting immunity following SARS-CoV-2 infection is questioned because serum antibodies decline in convalescence. However, functional immunity is mediated by long-lived memory T and B (Bmem) cells. Therefore, we generated fluorescently-labeled tetramers of the spike receptor binding domain (RBD) and nucleocapsid protein (NCP) to determine the longevity and immunophenotype of SARS-CoV-2-specific Bmem cells in COVID-19 patients. A total of 36 blood samples were obtained from 25 COVID-19 patients between 4 and 242 days post-symptom onset including 11 paired samples. While serum IgG to RBD and NCP was identified in all patients, antibody levels began declining at 20 days post-symptom onset. RBD- and NCP-specific Bmem cells predominantly expressed IgM+ or IgG1+ and continued to rise until 150 days. RBD-specific IgG+ Bmem were predominantly CD27+, and numbers significantly correlated with circulating follicular helper T cell numbers. Thus, the SARS-CoV-2 antibody response contracts in convalescence with persistence of RBD- and NCP-specific Bmem cells. Flow cytometric detection of SARS-CoV-2-specific Bmem cells enables detection of long-term immune memory following infection or vaccination for COVID-19.
Copyright © 2020, American Association for the Advancement of Science.
Figures


References
-
- Richardson S., Hirsch J. S., Narasimhan M., Crawford J. M., McGinn T., Davidson K. W., Barnaby D. P., Becker L. B., Chelico J. D., Cohen S. L., Cookingham J., Coppa K., Diefenbach M. A., Dominello A. J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T. G., Hirschwerk D. A., Kim E. J., Kozel Z. M., Marrast L. M., Mogavero J. N., Osorio G. A., Qiu M., Zanos T. P.; the Northwell COVID-19 Research Consortium , Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA 323, 2052–2059 (2020). 10.1001/jama.2020.6775 - DOI - PMC - PubMed
-
- Williamson E. J., Walker A. J., Bhaskaran K., Bacon S., Bates C., Morton C. E., Curtis H. J., Mehrkar A., Evans D., Inglesby P., Cockburn J., McDonald H. I., MacKenna B., Tomlinson L., Douglas I. J., Rentsch C. T., Mathur R., Wong A. Y. S., Grieve R., Harrison D., Forbes H., Schultze A., Croker R., Parry J., Hester F., Harper S., Perera R., Evans S. J. W., Smeeth L., Goldacre B., Factors associated with COVID-19-related death using OpenSAFELY. Nature 584, 430–436 (2020). 10.1038/s41586-020-2521-4 - DOI - PMC - PubMed
-
- Meyts I., Bucciol G., Quinti I., Neven B., Fischer A., Seoane E., Lopez-Granados E., Gianelli C., Robles-Marhuenda A., Jeandel P. Y., Paillard C., Sankaran V. G., Demirdag Y. Y., Lougaris V., Aiuti A., Plebani A., Milito C., Dalm V. A., Guevara-Hoyer K., Sánchez-Ramón S., Bezrodnik L., Barzaghi F., Gonzalez-Granado L. I., Hayman G. R., Uzel G., Mendonça L. O., Agostini C., Spadaro G., Badolato R., Soresina A., Vermeulen F., Bosteels C., Lambrecht B. N., Keller M., Mustillo P. J., Abraham R. S., Gupta S., Ozen A., Karakoc-Aydiner E., Baris S., Freeman A. F., Yamazaki-Nakashimada M., Scheffler-Mendoza S., Espinosa-Padilla S., Gennery A. R., Jolles S., Espinosa Y., Poli M. C., Fieschi C., Hauck F., Cunningham-Rundles C., Mahlaoui N., Warnatz K., Sullivan K. E., Tangye S. G.; IUIS Committee of Inborn Errors of Immunity , Coronavirus disease 2019 in patients with inborn errors of immunity: An international study. J. Allergy Clin. Immunol. S0091-6749(20)31320-8 (2020). 10.1016/j.jaci.2020.09.010 - DOI - PMC - PubMed
-
- Nguyen-Contant P., Embong A. K., Kanagaiah P., Chaves F. A., Yang H., Branche A. R., Topham D. J., Sangster M. Y., S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. mBio 11, e01991-20 (2020). 10.1128/mBio.01991-20 - DOI - PMC - PubMed
-
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). 10.1016/S0140-6736(20)30183-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

