Disrupting the DREAM transcriptional repressor complex induces apolipoprotein overexpression and systemic amyloidosis in mice
- PMID: 33444292
- PMCID: PMC7880409
- DOI: 10.1172/JCI140903
Disrupting the DREAM transcriptional repressor complex induces apolipoprotein overexpression and systemic amyloidosis in mice
Abstract
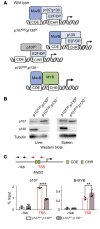
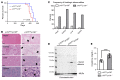
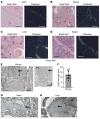
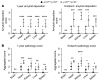
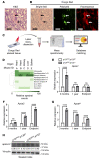
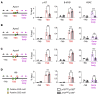
DREAM (Dp, Rb-like, E2F, and MuvB) is a transcriptional repressor complex that regulates cell proliferation, and its loss causes neonatal lethality in mice. To investigate DREAM function in adult mice, we used an assembly-defective p107 protein and conditional deletion of its redundant family member p130. In the absence of DREAM assembly, mice displayed shortened survival characterized by systemic amyloidosis but no evidence of excessive cellular proliferation. Amyloid deposits were found in the heart, liver, spleen, and kidneys but not the brain or bone marrow. Using laser-capture microdissection followed by mass spectrometry, we identified apolipoproteins as the most abundant components of amyloids. Intriguingly, apoA-IV was the most detected amyloidogenic protein in amyloid deposits, suggesting apoA-IV amyloidosis (AApoAIV). AApoAIV is a recently described form, whereby WT apoA-IV has been shown to predominate in amyloid plaques. We determined by ChIP that DREAM directly regulated Apoa4 and that the histone variant H2AZ was reduced from the Apoa4 gene body in DREAM's absence, leading to overexpression. Collectively, we describe a mechanism by which epigenetic misregulation causes apolipoprotein overexpression and amyloidosis, potentially explaining the origins of nongenetic amyloid subtypes.
Keywords: Epigenetics; Lipoproteins; Mouse models; Nephrology; Vascular Biology.
Conflict of interest statement
Figures

References
-
- Nuvolone M, Merlini G. Systemic amyloidosis: novel therapies and role of biomarkers. Nephrol Dial Transplant. 2017;32(5):770–780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

