Dispersal history of Miniopterus fuliginosus bats and their associated viruses in east Asia
- PMID: 33444317
- PMCID: PMC7808576
- DOI: 10.1371/journal.pone.0244006
Dispersal history of Miniopterus fuliginosus bats and their associated viruses in east Asia
Abstract
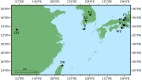
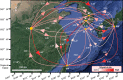
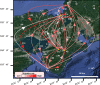
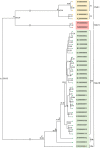
In this study, we examined the role of the eastern bent-winged bat (Miniopterus fuliginosus) in the dispersion of bat adenovirus and bat alphacoronavirus in east Asia, considering their gene flows and divergence times (based on deep-sequencing data), using bat fecal guano samples. Bats in China moved to Jeju Island and/or Taiwan in the last 20,000 years via the Korean Peninsula and/or Japan. The phylogenies of host mitochondrial D-loop DNA was not significantly congruent with those of bat adenovirus (m2XY = 0.07, p = 0.08), and bat alphacoronavirus (m2XY = 0.48, p = 0.20). We estimate that the first divergence time of bats carrying bat adenovirus in five caves studied (designated as K1, K2, JJ, N2, and F3) occurred approximately 3.17 million years ago. In contrast, the first divergence time of bat adenovirus among bats in the 5 caves was estimated to be approximately 224.32 years ago. The first divergence time of bats in caves CH, JJ, WY, N2, F1, F2, and F3 harboring bat alphacoronavirus was estimated to be 1.59 million years ago. The first divergence time of bat alphacoronavirus among the 7 caves was estimated to be approximately 2,596.92 years ago. The origin of bat adenovirus remains unclear, whereas our findings suggest that bat alphacoronavirus originated in Japan. Surprisingly, bat adenovirus and bat alphacoronavirus appeared to diverge substantially over the last 100 years, even though our gene-flow data indicate that the eastern bent-winged bat serves as an important natural reservoir of both viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The genetic diversity of D-loop sequences in eastern bent-winged bats (Miniopterus fuliginosus) living in Wakayama Prefecture, Japan.J Vet Med Sci. 2017 Jun 29;79(6):1142-1145. doi: 10.1292/jvms.17-0152. Epub 2017 May 8. J Vet Med Sci. 2017. PMID: 28484149 Free PMC article.
-
Full Genome of batCoV/MinFul/2018/SriLanka, a Novel Alpha-Coronavirus Detected in Miniopterus fuliginosus, Sri Lanka.Viruses. 2022 Feb 7;14(2):337. doi: 10.3390/v14020337. Viruses. 2022. PMID: 35215931 Free PMC article.
-
Identification of a new alphacoronavirus (Coronaviridae: Alphacoronavirus) associated with the greater horseshoe bat (Rhinolophus ferrumequinum) in the south of European part of Russia.Vopr Virusol. 2024 Dec 15;69(6):546-557. doi: 10.36233/0507-4088-279. Vopr Virusol. 2024. PMID: 39841419
-
Natural epigenetic variation in bats and its role in evolution.J Exp Biol. 2015 Jan 1;218(Pt 1):100-6. doi: 10.1242/jeb.107243. J Exp Biol. 2015. PMID: 25568456 Review.
-
Bat-Origin Coronaviruses Expand Their Host Range to Pigs.Trends Microbiol. 2018 Jun;26(6):466-470. doi: 10.1016/j.tim.2018.03.001. Epub 2018 Apr 18. Trends Microbiol. 2018. PMID: 29680361 Free PMC article. Review.
Cited by
-
Phylogeography of the Japanese greater horseshoe bat Rhinolophus nippon (Mammalia: Chiroptera) in Northeast Asia: New insight into the monophyly of the Japanese populations.Ecol Evol. 2021 Dec 3;11(24):18181-18195. doi: 10.1002/ece3.8414. eCollection 2021 Dec. Ecol Evol. 2021. PMID: 35003666 Free PMC article.
-
Increased urinary creatinine during hibernation and day roosting in the Eastern bent-winged bat (Miniopterus fuliginosus) in Korea.Commun Biol. 2024 Jan 5;7(1):42. doi: 10.1038/s42003-023-05713-1. Commun Biol. 2024. PMID: 38182741 Free PMC article.
-
Target enrichment metaviromics enables comprehensive surveillance of coronaviruses in environmental and animal samples.Heliyon. 2024 May 19;10(11):e31556. doi: 10.1016/j.heliyon.2024.e31556. eCollection 2024 Jun 15. Heliyon. 2024. PMID: 38845944 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

