Versatile CRISPR/Cas9-mediated mosaic analysis by gRNA-induced crossing-over for unmodified genomes
- PMID: 33444322
- PMCID: PMC7837743
- DOI: 10.1371/journal.pbio.3001061
Versatile CRISPR/Cas9-mediated mosaic analysis by gRNA-induced crossing-over for unmodified genomes
Abstract
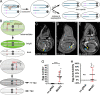
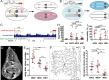
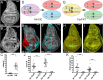
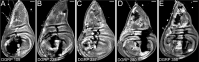
Mosaic animals have provided the platform for many fundamental discoveries in developmental biology, cell biology, and other fields. Techniques to produce mosaic animals by mitotic recombination have been extensively developed in Drosophila melanogaster but are less common for other laboratory organisms. Here, we report mosaic analysis by gRNA-induced crossing-over (MAGIC), a new technique for generating mosaic animals based on DNA double-strand breaks produced by CRISPR/Cas9. MAGIC efficiently produces mosaic clones in both somatic tissues and the germline of Drosophila. Further, by developing a MAGIC toolkit for 1 chromosome arm, we demonstrate the method's application in characterizing gene function in neural development and in generating fluorescently marked clones in wild-derived Drosophila strains. Eliminating the need to introduce recombinase-recognition sites into the genome, this simple and versatile system simplifies mosaic analysis in Drosophila and can in principle be applied in any organism that is compatible with CRISPR/Cas9.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

