UV-exposure, endogenous DNA damage, and DNA replication errors shape the spectra of genome changes in human skin
- PMID: 33444353
- PMCID: PMC7808690
- DOI: 10.1371/journal.pgen.1009302
UV-exposure, endogenous DNA damage, and DNA replication errors shape the spectra of genome changes in human skin
Abstract
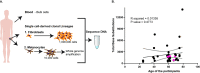
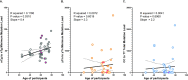
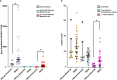
Human skin is continuously exposed to environmental DNA damage leading to the accumulation of somatic mutations over the lifetime of an individual. Mutagenesis in human skin cells can be also caused by endogenous DNA damage and by DNA replication errors. The contributions of these processes to the somatic mutation load in the skin of healthy humans has so far not been accurately assessed because the low numbers of mutations from current sequencing methodologies preclude the distinction between sequencing errors and true somatic genome changes. In this work, we sequenced genomes of single cell-derived clonal lineages obtained from primary skin cells of a large cohort of healthy individuals across a wide range of ages. We report here the range of mutation load and a comprehensive view of the various somatic genome changes that accumulate in skin cells. We demonstrate that UV-induced base substitutions, insertions and deletions are prominent even in sun-shielded skin. In addition, we detect accumulation of mutations due to spontaneous deamination of methylated cytosines as well as insertions and deletions characteristic of DNA replication errors in these cells. The endogenously induced somatic mutations and indels also demonstrate a linear increase with age, while UV-induced mutation load is age-independent. Finally, we show that DNA replication stalling at common fragile sites are potent sources of gross chromosomal rearrangements in human cells. Thus, somatic mutations in skin of healthy individuals reflect the interplay of environmental and endogenous factors in facilitating genome instability and carcinogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Abyzov A, Tomasini L, Zhou B, Vasmatzis N, Coppola G, Amenduni M, et al. One thousand somatic SNVs per skin fibroblast cell set baseline of mosaic mutational load with patterns that suggest proliferative origin. Genome Res. 2017;27(4):512–23. Epub 2017/02/24. 10.1101/gr.215517.116 . - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

