The complex architecture of p53 binding sites
- PMID: 33444431
- PMCID: PMC7897521
- DOI: 10.1093/nar/gkaa1283
The complex architecture of p53 binding sites
Abstract
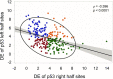
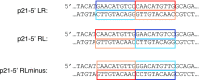
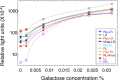
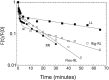
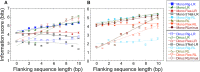
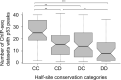
Sequence-specific protein-DNA interactions are at the heart of the response of the tumor-suppressor p53 to numerous physiological and stress-related signals. Large variability has been previously reported in p53 binding to and transactivating from p53 response elements (REs) due, at least in part, to changes in direct (base) and indirect (shape) readouts of p53 REs. Here, we dissect p53 REs to decipher the mechanism by which p53 optimizes this highly regulated variable level of interaction with its DNA binding sites. We show that hemi-specific binding is more prevalent in p53 REs than previously envisioned. We reveal that sequences flanking the REs modulate p53 binding and activity and show that these effects extend to 4-5 bp from the REs. Moreover, we show here that the arrangement of p53 half-sites within its REs, relative to transcription direction, has been fine-tuned by selection pressure to optimize and regulate the response levels from p53 REs. This directionality in the REs arrangement is at least partly encoded in the structural properties of the REs. Furthermore, we show here that in the p21-5' RE the orientation of the half-sites is such that the effect of the flanking sequences is minimized and we discuss its advantages.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Kitayner M., Rozenberg H., Kessler N., Rabinovich D., Shaulov L., Haran T.E., Shakked Z.. Structural basis of DNA recognition by p53 tetramers. Mol. Cell. 2006; 22:741–753. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

