scRNA-Seq Reveals New Enteric Nervous System Roles for GDNF, NRTN, and TBX3
- PMID: 33444816
- PMCID: PMC8099699
- DOI: 10.1016/j.jcmgh.2020.12.014
scRNA-Seq Reveals New Enteric Nervous System Roles for GDNF, NRTN, and TBX3
Abstract
Background and aims: Bowel function requires coordinated activity of diverse enteric neuron subtypes. Our aim was to define gene expression in these neuron subtypes to facilitate development of novel therapeutic approaches to treat devastating enteric neuropathies, and to learn more about enteric nervous system function.
Methods: To identify subtype-specific genes, we performed single-nucleus RNA-seq on adult mouse and human colon myenteric plexus, and single-cell RNA-seq on E17.5 mouse ENS cells from whole bowel. We used immunohistochemistry, select mutant mice, and calcium imaging to validate and extend results.
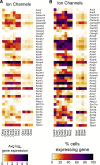
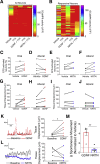
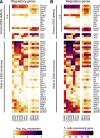
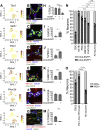
Results: RNA-seq on 635 adult mouse colon myenteric neurons and 707 E17.5 neurons from whole bowel defined seven adult neuron subtypes, eight E17.5 neuron subtypes and hundreds of differentially expressed genes. Manually dissected human colon myenteric plexus yielded RNA-seq data from 48 neurons, 3798 glia, 5568 smooth muscle, 377 interstitial cells of Cajal, and 2153 macrophages. Immunohistochemistry demonstrated differential expression for BNC2, PBX3, SATB1, RBFOX1, TBX2, and TBX3 in enteric neuron subtypes. Conditional Tbx3 loss reduced NOS1-expressing myenteric neurons. Differential Gfra1 and Gfra2 expression coupled with calcium imaging revealed that GDNF and neurturin acutely and differentially regulate activity of ∼50% of myenteric neurons with distinct effects on smooth muscle contractions.
Conclusion: Single cell analyses defined genes differentially expressed in myenteric neuron subtypes and new roles for TBX3, GDNF and NRTN. These data facilitate molecular diagnostic studies and novel therapeutics for bowel motility disorders.
Keywords: Calcium Imaging; Human and Mouse Colon; Pou3f3 (Brn1); Transcription Factors.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures














References
-
- Schneider S., Wright C.M., Heuckeroth R.O. Unexpected roles for the second brain: enteric nervous system as master regulator of bowel function. Annual Review of Physiology. 2019;81:235–259. - PubMed
-
- McCann C.J., Thapar N. Enteric neural stem cell therapies for enteric neuropathies. Neurogastroenterol Motil. 2018;30:e13369. - PubMed
-
- Soret R., Schneider S., Bernas G., Christophers B., Souchkova O., Charrier B., Righini-Grunder F., Aspirot A., Landry M., Kembel S.W., Faure C., Heuckeroth R.O., Pilon N. Glial cell derived neurotrophic factor induces enteric neurogenesis and improves colon structure and function in mouse models of Hirschsprung disease. Gastroenterology. 2020;159:1824–1838.e7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

