Protein N-glycosylation and O-mannosylation are catalyzed by two evolutionarily related GT-C glycosyltransferases
- PMID: 33445129
- PMCID: PMC8222153
- DOI: 10.1016/j.sbi.2020.12.009
Protein N-glycosylation and O-mannosylation are catalyzed by two evolutionarily related GT-C glycosyltransferases
Abstract
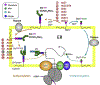
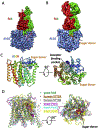
The structural folds of glycosyltransferases are categorized into three superfamilies: GT-A, GT-B, and GT-C. Few structures of GT-C fold existed in the Protein Data Bank prior to the recent advent of high-resolution cryo-EM, because the glycosyltransferases are large membrane proteins that are difficult to crystallize. The use of cryo-EM has resulted in the structures of several key GT-C glycosyltransferases. Here we summarize the latest structural features of and mechanistic insights into these membrane enzyme complexes.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of interest statement
Nothing declared.
Figures


References
-
- Helenius A, Aebi M: Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 2004, 73:1019–1049. - PubMed
-
- Lairson LL, Henrissat B, Davies GJ, Withers SG: Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 2008, 77:521–555. - PubMed
-
- Spiro RG: Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002, 12:43r–56r. - PubMed
-
- Bai XC, McMullan G, Scheres SH: How cryo-EM is revolutionizing structural biology. Trends Biochem Sci 2015, 40:49–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

