Inferring recombination patterns in African populations
- PMID: 33445180
- PMCID: PMC8117444
- DOI: 10.1093/hmg/ddab020
Inferring recombination patterns in African populations
Abstract
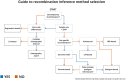
Although several high-resolution recombination maps exist for European-descent populations, the recombination landscape of African populations remains relatively understudied. Given that there is high genetic divergence among groups in Africa, it is possible that recombination hotspots also diverge significantly. Both limitations and opportunities exist for developing recombination maps for these populations. In this review, we discuss various recombination inference methods, and the strengths and weaknesses of these methods in analyzing recombination in African-descent populations. Furthermore, we provide a decision tree and recommendations for which inference method to use in various research contexts. Establishing an appropriate methodology for recombination rate inference in a particular study will improve the accuracy of various downstream analyses including but not limited to local ancestry inference, haplotype phasing, fine-mapping of GWAS loci and genome assemblies.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
Similar articles
-
Genome-wide Ancestry and Demographic History of African-Descendant Maroon Communities from French Guiana and Suriname.Am J Hum Genet. 2017 Nov 2;101(5):725-736. doi: 10.1016/j.ajhg.2017.09.021. Am J Hum Genet. 2017. PMID: 29100086 Free PMC article.
-
Similarity in recombination rate and linkage disequilibrium at CYP2C and CYP2D cytochrome P450 gene regions among Europeans indicates signs of selection and no advantage of using tagSNPs in population isolates.Pharmacogenet Genomics. 2012 Dec;22(12):846-57. doi: 10.1097/FPC.0b013e32835a3a6d. Pharmacogenet Genomics. 2012. PMID: 23089684
-
The critical needs and challenges for genetic architecture studies in Africa.Curr Opin Genet Dev. 2018 Dec;53:113-120. doi: 10.1016/j.gde.2018.08.005. Epub 2018 Sep 18. Curr Opin Genet Dev. 2018. PMID: 30240950 Free PMC article. Review.
-
The genomic landscape of African populations in health and disease.Hum Mol Genet. 2017 Oct 1;26(R2):R225-R236. doi: 10.1093/hmg/ddx253. Hum Mol Genet. 2017. PMID: 28977439 Free PMC article. Review.
-
Genome-wide fine-scale recombination rate variation in Drosophila melanogaster.PLoS Genet. 2012;8(12):e1003090. doi: 10.1371/journal.pgen.1003090. Epub 2012 Dec 20. PLoS Genet. 2012. PMID: 23284288 Free PMC article.
Cited by
-
Exploring Meiotic Recombination and Its Potential Benefits in South African Beef Cattle: A Review.Vet Sci. 2025 Jul 16;12(7):669. doi: 10.3390/vetsci12070669. Vet Sci. 2025. PMID: 40711329 Free PMC article. Review.
-
Fine human genetic map based on UK10K data set.Hum Genet. 2022 Feb;141(2):273-281. doi: 10.1007/s00439-021-02415-8. Epub 2022 Jan 20. Hum Genet. 2022. PMID: 35048190
-
Variation in fine-scale recombination rate in temperature-evolved Drosophila melanogaster populations in response to selection.G3 (Bethesda). 2022 Sep 30;12(10):jkac208. doi: 10.1093/g3journal/jkac208. G3 (Bethesda). 2022. PMID: 35961026 Free PMC article.
-
The recombination landscape of the Khoe-San likely represents the upper limits of recombination divergence in humans.Genome Biol. 2022 Aug 9;23(1):172. doi: 10.1186/s13059-022-02744-5. Genome Biol. 2022. PMID: 35945619 Free PMC article.
References
-
- Hunter, N. (2007) Meiotic recombination. In Aguilera, A. and Rothstein, R. (eds), Molecular Genetics of Recombination. Springer Berlin Heidelberg, Berlin, Heidelberg, pp. 381–442.
-
- Bergero, R. and Charlesworth, D. (2009) The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol (Amst), 24, 94–102. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

