Inferring recombination patterns in African populations
- PMID: 33445180
- PMCID: PMC8117444
- DOI: 10.1093/hmg/ddab020
Inferring recombination patterns in African populations
Abstract
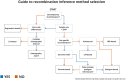
Although several high-resolution recombination maps exist for European-descent populations, the recombination landscape of African populations remains relatively understudied. Given that there is high genetic divergence among groups in Africa, it is possible that recombination hotspots also diverge significantly. Both limitations and opportunities exist for developing recombination maps for these populations. In this review, we discuss various recombination inference methods, and the strengths and weaknesses of these methods in analyzing recombination in African-descent populations. Furthermore, we provide a decision tree and recommendations for which inference method to use in various research contexts. Establishing an appropriate methodology for recombination rate inference in a particular study will improve the accuracy of various downstream analyses including but not limited to local ancestry inference, haplotype phasing, fine-mapping of GWAS loci and genome assemblies.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Hunter, N. (2007) Meiotic recombination. In Aguilera, A. and Rothstein, R. (eds), Molecular Genetics of Recombination. Springer Berlin Heidelberg, Berlin, Heidelberg, pp. 381–442.
-
- Bergero, R. and Charlesworth, D. (2009) The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol (Amst), 24, 94–102. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

