Identification of a New HIV-1 BC Intersubtype Circulating Recombinant Form (CRF108_BC) in Spain
- PMID: 33445523
- PMCID: PMC7826730
- DOI: 10.3390/v13010093
Identification of a New HIV-1 BC Intersubtype Circulating Recombinant Form (CRF108_BC) in Spain
Abstract
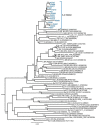
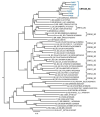
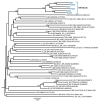
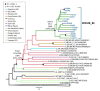
The extraordinary genetic variability of human immunodeficiency virus type 1 (HIV-1) group M has led to the identification of 10 subtypes, 102 circulating recombinant forms (CRFs) and numerous unique recombinant forms. Among CRFs, 11 derived from subtypes B and C have been identified in China, Brazil, and Italy. Here we identify a new HIV-1 CRF_BC in Northern Spain. Originally, a phylogenetic cluster of 15 viruses of subtype C in protease-reverse transcriptase was identified in an HIV-1 molecular surveillance study in Spain, most of them from individuals from the Basque Country and heterosexually transmitted. Analyses of near full-length genome sequences from six viruses from three cities revealed that they were BC recombinant with coincident mosaic structures different from known CRFs. This allowed the definition of a new HIV-1 CRF designated CRF108_BC, whose genome is predominantly of subtype C, with four short subtype B fragments. Phylogenetic analyses with database sequences supported a Brazilian ancestry of the parental subtype C strain. Coalescent Bayesian analyses estimated the most recent common ancestor of CRF108_BC in the city of Vitoria, Basque Country, around 2000. CRF108_BC is the first CRF_BC identified in Spain and the second in Europe, after CRF60_BC, both phylogenetically related to Brazilian subtype C strains.
Keywords: HIV-1; HIV-1 genetic diversity; HIV-1 molecular epidemiology; HIV-1 phylogeny; circulating recombinant form.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Molecular analysis of multiple HIV-1 BC recombinant strains circulating worldwide shows predominance of the C genotype in the viral genomic structure.Braz J Microbiol. 2025 Jun;56(2):883-892. doi: 10.1007/s42770-025-01629-6. Epub 2025 Apr 16. Braz J Microbiol. 2025. PMID: 40237921
-
Identification of a new HIV type 1 circulating BF intersubtype recombinant form (CRF47_BF) in Spain.AIDS Res Hum Retroviruses. 2010 Jul;26(7):827-32. doi: 10.1089/aid.2009.0311. AIDS Res Hum Retroviruses. 2010. PMID: 20618102
-
Diversity of mosaic structures and common ancestry of human immunodeficiency virus type 1 BF intersubtype recombinant viruses from Argentina revealed by analysis of near full-length genome sequences.J Gen Virol. 2002 Jan;83(Pt 1):107-119. doi: 10.1099/0022-1317-83-1-107. J Gen Virol. 2002. PMID: 11752707
-
Country Level Diversity of the HIV-1 Pandemic between 1990 and 2015.J Virol. 2020 Dec 22;95(2):e01580-20. doi: 10.1128/JVI.01580-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33087461 Free PMC article.
-
HIV-1 subtype frequency in Northeast Brazil: A systematic review and meta-analysis.J Med Virol. 2020 Dec;92(12):3219-3229. doi: 10.1002/jmv.25842. Epub 2020 Apr 28. J Med Virol. 2020. PMID: 32266997
Cited by
-
Molecular analysis of multiple HIV-1 BC recombinant strains circulating worldwide shows predominance of the C genotype in the viral genomic structure.Braz J Microbiol. 2025 Jun;56(2):883-892. doi: 10.1007/s42770-025-01629-6. Epub 2025 Apr 16. Braz J Microbiol. 2025. PMID: 40237921
-
Viruses Previously Identified in Brazil as Belonging to HIV-1 CRF72_BF1 Represent Two Closely Related Circulating Recombinant Forms, One of Which, Designated CRF122_BF1, Is Also Circulating in Spain.Front Microbiol. 2022 May 27;13:863084. doi: 10.3389/fmicb.2022.863084. eCollection 2022. Front Microbiol. 2022. PMID: 35694315 Free PMC article.
-
Nanopore Sequencing for Characterization of HIV-1 Recombinant Forms.Microbiol Spectr. 2022 Aug 31;10(4):e0150722. doi: 10.1128/spectrum.01507-22. Epub 2022 Jul 27. Microbiol Spectr. 2022. PMID: 35894615 Free PMC article.
-
Epidemiological and molecular characterization of HBV and HCV infections in HIV-1-infected inmate population in Italy: a 2017-2019 multicenter cross-sectional study.Sci Rep. 2023 Sep 9;13(1):14908. doi: 10.1038/s41598-023-41814-x. Sci Rep. 2023. PMID: 37689795 Free PMC article.
-
Identification of a HIV-1 circulating BF1 recombinant form (CRF75_BF1) of Brazilian origin that also circulates in Southwestern Europe.Front Microbiol. 2023 Nov 30;14:1301374. doi: 10.3389/fmicb.2023.1301374. eCollection 2023. Front Microbiol. 2023. PMID: 38125564 Free PMC article.
References
-
- Gryseels S., Watts T.D., Mpolesha J.M.K., Larsen B.B., Lemey P., Muyembe-Tamfum J.J., Teuwen D.E., Worobey M. A near full-length HIV-1 genome from 1966 recovered from formalin-fixed paraffin-embedded tissue. Proc. Natl. Acad. Sci. USA. 2020;117:12222–12229. doi: 10.1073/pnas.1913682117. - DOI - PMC - PubMed
-
- Hemelaar J., Elangovan R., Yun J., Dickson-Tetteh L., Fleminger I., Kirtley S., Williams B., Gouws-Williams E., Ghys P.D., Abimiku A.G., et al. Global and regional molecular epidemiology of HIV-1, 1990–2015: A systematic review, global survey, and trend analysis. Lancet Infect. Dis. 2019;19:143–155. doi: 10.1016/S1473-3099(18)30647-9. - DOI - PubMed
-
- Hofstra L.M., Sauvageot N., Albert J., Alexiev I., García F., Struck D., Van De Vijver D.A.M.C., Åsjö B., Kolupajeva T., Kostrikis L.G., et al. Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin. Infect. Dis. 2016;62:655–663. doi: 10.1093/cid/civ963. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

