SARS-CoV-2 in Danish Mink Farms: Course of the Epidemic and a Descriptive Analysis of the Outbreaks in 2020
- PMID: 33445704
- PMCID: PMC7828158
- DOI: 10.3390/ani11010164
SARS-CoV-2 in Danish Mink Farms: Course of the Epidemic and a Descriptive Analysis of the Outbreaks in 2020
Abstract
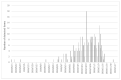
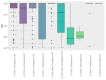
SARS-CoV-2 infection is the cause of COVID-19 in humans. In April 2020, SARS-CoV-2 infection in farmed mink (Neovision vision) occurred in the Netherlands. The first outbreaks in Denmark were detected in June 2020 in three farms. A steep increase in the number of infected farms occurred from September and onwards. Here, we describe prevalence data collected from 215 infected mink farms to characterize spread and impact of disease in infected farms. In one third of the farms, no clinical signs were observed. In farms with clinical signs, decreased feed intake, increased mortality and respiratory symptoms were most frequently observed, during a limited time period (median of 11 days). In 65% and 69% of farms, virus and sero-conversion, respectively, were detected in 100% of sampled animals at the first sampling. SARS-CoV-2 was detected, at low levels, in air samples collected close to the mink, on mink fur, on flies, on the foot of a seagull, and in gutter water, but not in feed. Some dogs and cats from infected farms tested positive for the virus. Chickens, rabbits, and horses sampled on a few farms, and wildlife sampled in the vicinity of the infected farms did not test positive for SARS-CoV-2. Thus, mink are highly susceptible to infection by SARS-CoV-2, but routes of transmission between farms, other than by direct human contact, are unclear.
Keywords: COVID-19, increased mortality; Neovision vision; SARS-CoV-2 prevalence; clinical signs; environment; seroprevalence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

