The Long-Term Evolutionary History of Gradual Reduction of CpG Dinucleotides in the SARS-CoV-2 Lineage
- PMID: 33445785
- PMCID: PMC7828247
- DOI: 10.3390/biology10010052
The Long-Term Evolutionary History of Gradual Reduction of CpG Dinucleotides in the SARS-CoV-2 Lineage
Abstract
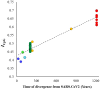
Recent studies suggested that the fraction of CG dinucleotides (CpG) is severely reduced in the genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The CpG deficiency was predicted to be the adaptive response of the virus to evade degradation of the viral RNA by the antiviral zinc finger protein that specifically binds to CpG nucleotides. By comparing all representative genomes belonging to the genus Betacoronavirus, this study examined the potential time of origin of CpG depletion. The results of this investigation revealed a highly significant correlation between the proportions of CpG nucleotide (CpG content) of the betacoronavirus species and their times of divergence from SARS-CoV-2. Species that are distantly related to SARS-CoV-2 had much higher CpG contents than that of SARS-CoV-2. Conversely, closely related species had low CpG contents that are similar to or slightly higher than that of SARS-CoV-2. These results suggest a systematic and continuous reduction in the CpG content in the SARS-CoV-2 lineage that might have started since the Sarbecovirus + Hibecovirus clade separated from Nobecovirus, which was estimated to be 1213 years ago. This depletion was not found to be mediated by the GC contents of the genomes. Our results also showed that the depletion of CpG occurred at neutral positions of the genome as well as those under selection. The latter is evident from the progressive reduction in the proportion of arginine amino acid (coded by CpG dinucleotides) in the SARS-CoV-2 lineage over time. The results of this study suggest that shedding CpG nucleotides from their genome is a continuing process in this viral lineage, potentially to escape from their host defense mechanisms.
Keywords: COVID-19; CpG dinucleotide; SARS-CoV-2; Sarbecovirus; adaptation; host defense; virus evolution; zinc finger protein.
Conflict of interest statement
The author declares no conflict of interest.
Figures

Similar articles
-
SARS-CoV-2 within-host diversity of human hosts and its implications for viral immune evasion.mBio. 2023 Aug 31;14(4):e0067923. doi: 10.1128/mbio.00679-23. Epub 2023 Jun 5. mBio. 2023. PMID: 37273216 Free PMC article.
-
The Slowing Rate of CpG Depletion in SARS-CoV-2 Genomes Is Consistent with Adaptations to the Human Host.Mol Biol Evol. 2022 Mar 2;39(3):msac029. doi: 10.1093/molbev/msac029. Mol Biol Evol. 2022. PMID: 35134218 Free PMC article.
-
Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2.J Med Virol. 2020 Jun;92(6):602-611. doi: 10.1002/jmv.25731. Epub 2020 Mar 11. J Med Virol. 2020. PMID: 32104911 Free PMC article.
-
SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus-Host Interaction.Int J Mol Sci. 2020 Jun 26;21(12):4549. doi: 10.3390/ijms21124549. Int J Mol Sci. 2020. PMID: 32604730 Free PMC article. Review.
-
A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses.Emerg Microbes Infect. 2020 Mar 14;9(1):558-570. doi: 10.1080/22221751.2020.1736644. eCollection 2020. Emerg Microbes Infect. 2020. PMID: 32172672 Free PMC article. Review.
Cited by
-
The low abundance of CpG in the SARS-CoV-2 genome is not an evolutionarily signature of ZAP.Sci Rep. 2022 Feb 14;12(1):2420. doi: 10.1038/s41598-022-06046-5. Sci Rep. 2022. PMID: 35165300 Free PMC article.
-
Natural selection plays a significant role in governing the codon usage bias in the novel SARS-CoV-2 variants of concern (VOC).PeerJ. 2022 Jun 23;10:e13562. doi: 10.7717/peerj.13562. eCollection 2022. PeerJ. 2022. PMID: 35765592 Free PMC article.
-
Overview of host-directed antiviral targets for future research and drug development.Acta Pharm Sin B. 2025 Apr;15(4):1723-1751. doi: 10.1016/j.apsb.2025.03.011. Epub 2025 Mar 8. Acta Pharm Sin B. 2025. PMID: 40486850 Free PMC article. Review.
-
Pulmonary Delivery of Nonviral Nucleic Acid-Based Vaccines With Spotlight on Gold Nanoparticles.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2025 Jan-Feb;17(1):e70000. doi: 10.1002/wnan.70000. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2025. PMID: 39800783 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

