The first detection of Anaplasma capra, an emerging zoonotic Anaplasma sp., in erythrocytes
- PMID: 33446064
- PMCID: PMC7894429
- DOI: 10.1080/22221751.2021.1876532
The first detection of Anaplasma capra, an emerging zoonotic Anaplasma sp., in erythrocytes
Abstract
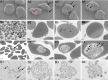
ABSTRACT An emerging infectious disease caused by "Anaplasma capra" was reported in a 2015 survey of 477 hospital patients with a tick-bite history in China. However, the morphological characteristics and parasitic location of this pathogen are still unclear, and the pathogen has not been officially classified as a member of the genus Anaplasma. Anaplasma capra-positive blood samples were collected, blood cells separated, and DNA of whole blood cells, erythrocytes, and leukocytes extracted. Multiplex PCR detection assay was used to detect whole blood cell, erythrocytes and leukocytes, DNA samples, and PCR identification, nucleic acid sequencing, and phylogenetic analyses based on A. capra groEL, 16S rRNA, gltA, and msp4 genes. Scanning electron microscopy (SEM), transmission electron microscopy (TEM), Wright-Giemsa staining, chromogenic in situ hybridization (CISH), immunocytochemistry, and indirect immunofluorescence assay (IFA) were used to identify the location and morphological characteristics of A. capra. Multiple gene loci results demonstrated that erythrocyte DNA samples were A. capra-positive, while leukocyte DNA samples were A. capra-negative. Phylogenetic analysis showed that A. capra is in the same clade with the A. capra sequence reported previously. SEM and TEM showed one or more pathogens internally or on the outer surface of erythrocytes. Giemsa staining, CISH, immunocytochemistry, and IFA indicated that erythrocytes were A. capra-positive. This study is the first to identify the novel zoonotic tick-borne Anaplasma sp., "Anaplasma capra," in host erythrocytes. Based on our results, we suggest revision of Genus Anaplasma and formally name "A. capra" as Anaplasma capra sp. nov.
Keywords: Anaplasma capra; Rickettsia; erythrocyte; tick-borne; zoonotic.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
-
- Li H, Zheng YC, Ma L, et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect Dis. 2015;15(6):663–670. - PubMed
-
- Zhou Z, Nie K, Tang C, et al. Phylogenetic analysis of the genus Anaplasma in Southwestern China based on 16S rRNA sequence. Res Vet Sci. 2010;89(2):262–265. - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
