Novel application of single-cell next-generation sequencing for determination of intratumoral heterogeneity of canine osteosarcoma cell lines
- PMID: 33446089
- PMCID: PMC7944434
- DOI: 10.1177/1040638720985242
Novel application of single-cell next-generation sequencing for determination of intratumoral heterogeneity of canine osteosarcoma cell lines
Abstract
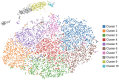
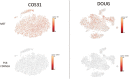
Osteosarcoma (OSA) is a highly aggressive and metastatic neoplasm of both the canine and human patient and is the leading form of osseous neoplasia in both species worldwide. To gain deeper insight into the heterogeneous and genetically chaotic nature of OSA, we applied single-cell transcriptome (scRNA-seq) analysis to 4 canine OSA cell lines. This novel application of scRNA-seq technology to the canine genome required uploading the CanFam3.1 reference genome into an analysis pipeline (10X Genomics Cell Ranger); this methodology has not been reported previously in the canine species, to our knowledge. The scRNA-seq outputs were validated by comparing them to cDNA expression from reverse-transcription PCR (RT-PCR) and Sanger sequencing bulk analysis of 4 canine OSA cell lines (COS31, DOUG, POS, and HMPOS) for 11 genes implicated in the pathogenesis of canine OSA. The scRNA-seq outputs revealed the significant heterogeneity of gene transcription expression patterns within the cell lines investigated (COS31 and DOUG). The scRNA-seq data showed 10 distinct clusters of similarly shared transcriptomic expression patterns in COS31; 12 clusters were identified in DOUG. In addition, cRNA-seq analysis provided data for integration into the Qiagen Ingenuity Pathway Analysis software for canonical pathway analysis. Of the 81 distinct pathways identified within the clusters, 33 had been implicated in the pathogenesis of OSA, of which 18 had not been reported previously in canine OSA.
Keywords: canine; neoplasia; osteosarcoma; single-cell transcriptomics; tumor heterogeneity.
Conflict of interest statement
Figures


References
-
- Bao H, et al. Potential mechanisms underlying CDK5 related osteosarcoma progression. Expert Opin Ther Targets 2017;21:455–460. - PubMed
-
- Barroga EF, et al. Establishment and characterization of the growth and pulmonary metastasis of a highly lung metastasizing cell line from canine osteosarcoma in nude mice. J Vet Med Sci 1999;61:361–367. - PubMed
-
- Baslan T, Hicks J. Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nature Rev Cancer 2017;17:557–569. - PubMed
-
- Bergman PJ, et al. Amputation and carboplatin for treatment of dogs with osteosarcoma: 48 cases (1991 to 1993). J Vet Intern Med 1996;10:76–81. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

