Genomic regions associated with stripe rust resistance against the Egyptian race revealed by genome-wide association study
- PMID: 33446120
- PMCID: PMC7809828
- DOI: 10.1186/s12870-020-02813-6
Genomic regions associated with stripe rust resistance against the Egyptian race revealed by genome-wide association study
Abstract
Background: Wheat stripe rust (caused by Puccinia striiformis f. sp. Tritici), is a major disease that causes huge yield damage. New pathogen races appeared in the last few years and caused a broke down in the resistant genotypes. In Egypt, some of the resistant genotypes began to be susceptible to stripe rust in recent years. This situation increases the need to produce new genotypes with durable resistance. Besides, looking for a new resistant source from the available wheat genotypes all over the world help in enhancing the breeding programs.
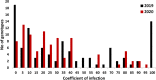
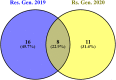
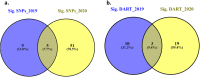
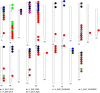
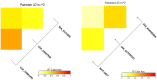
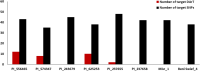
Results: In the recent study, a set of 103-spring wheat genotypes from different fourteen countries were evaluated to their field resistant to stripe rust for two years. These genotypes included 17 Egyptian genotypes from the old and new cultivars. The 103-spring wheat genotypes were reported to be well adapted to the Egyptian environmental conditions. Out of the tested genotypes, eight genotypes from four different countries were found to be resistant in both years. Genotyping was carried out using genotyping-by-sequencing and a set of 26,703 SNPs were used in the genome-wide association study. Five SNP markers, located on chromosomes 2A and 4A, were found to be significantly associated with the resistance in both years. Three gene models associated with disease resistance and underlying these significant SNPs were identified. One immune Iranian genotype, with the highest number of different alleles from the most resistant Egyptian genotypes, was detected.
Conclusion: the high variation among the tested genotypes in their resistance to the Egyptian stripe rust race confirming the possible improvement of stripe rust resistance in the Egyptian wheat genotypes. The identified five SNP markers are stable and could be used in marker-assisted selection after validation in different genetic backgrounds. Crossing between the immune Iranian genotype and the Egyptian genotypes will improve stripe rust resistance in Egypt.
Keywords: Coefficient of Infection; Disease severity; Genome-wide association study; Linkage disequilibrium; Single marker analysis; gene expression.
Conflict of interest statement
The authors declare that they have no competing interests
Figures

References
-
- Wellings CR. Global status of stripe rust: A review of historical and current threats. Euphytica. 2011;179:129–141. doi: 10.1007/s10681-011-0360-y. - DOI
-
- Chen XM. Epidemiology and control of stripe rust Puccinia striiformis f . sp . tritici on wheat. Can J Plant Pathol. 2005;27:314–337. doi: 10.1080/07060660509507230. - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

