Deciphering molecular mechanisms underlying chemoresistance in relapsed AML patients: towards precision medicine overcoming drug resistance
- PMID: 33446189
- PMCID: PMC7809753
- DOI: 10.1186/s12935-021-01746-w
Deciphering molecular mechanisms underlying chemoresistance in relapsed AML patients: towards precision medicine overcoming drug resistance
Abstract
Background: Acute myeloid leukemia (AML) remains a devastating disease with a 5-year survival rate of less than 30%. AML treatment has undergone significant changes in recent years, incorporating novel targeted therapies along with improvements in allogeneic bone marrow transplantation techniques. However, the standard of care remains cytarabine and anthracyclines, and the primary hindrance towards curative treatment is the frequent emergence of intrinsic and acquired anticancer drug resistance. In this respect, patients presenting with chemoresistant AML face dismal prognosis even with most advanced therapies. Herein, we aimed to explore the potential implementation of the characterization of chemoresistance mechanisms in individual AML patients towards efficacious personalized medicine.
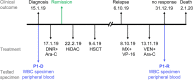
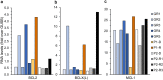
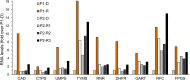
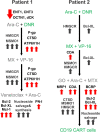
Methods: Towards the identification of tailored treatments for individual patients, we herein present the cases of relapsed AML patients, and compare them to patients displaying durable remissions following the same chemotherapeutic induction treatment. We quantified the expression levels of specific genes mediating drug transport and metabolism, nucleotide biosynthesis, and apoptosis, in order to decipher the molecular mechanisms underlying intrinsic and/or acquired chemoresistance modalities in relapsed patients. This was achieved by real-time PCR using patient cDNA, and could be readily implemented in the clinical setting.
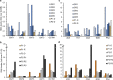
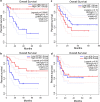
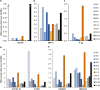
Results: This analysis revealed pre-existing differences in gene expression levels between the relapsed patients and patients with lasting remissions, as well as drug-induced alterations at different relapse stages compared to diagnosis. Each of the relapsed patients displayed unique chemoresistance mechanisms following similar treatment protocols, which could have been missed in a large study aimed at identifying common drug resistance determinants.
Conclusions: Our findings emphasize the need for standardized evaluation of key drug transport and metabolism genes as an integral component of routine AML management, thereby allowing for the selection of treatments of choice for individual patients. This approach could facilitate the design of efficacious personalized treatment regimens, thereby reducing relapse rates of therapy refractory disease.
Keywords: AML; Chemotherapy; Drug metabolism; Intrinsic/acquired chemoresistance; Precision medicine; Relapse; Resistance modalities.
Conflict of interest statement
The authors declare that they have no competing interests
Figures

References
-
- Cancer Statistics Review, 1975–2017—SEER Statistics. https://seer.cancer.gov/csr/1975_2017/. Accessed 18 Jul 2020.
LinkOut - more resources
Full Text Sources
Other Literature Sources

