Different methylation signatures at diagnosis in patients with high-risk myelodysplastic syndromes and secondary acute myeloid leukemia predict azacitidine response and longer survival
- PMID: 33446256
- PMCID: PMC7809812
- DOI: 10.1186/s13148-021-01002-y
Different methylation signatures at diagnosis in patients with high-risk myelodysplastic syndromes and secondary acute myeloid leukemia predict azacitidine response and longer survival
Abstract
Background: Epigenetic therapy, using hypomethylating agents (HMA), is known to be effective in the treatment of high-risk myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) patients who are not suitable for intensive chemotherapy and/or allogeneic stem cell transplantation. However, response rates to HMA are low and there is an unmet need in finding prognostic and predictive biomarkers of treatment response and overall survival. We performed global methylation analysis of 75 patients with high-risk MDS and secondary AML who were included in CETLAM SMD-09 protocol, in which patients received HMA or intensive treatment according to age, comorbidities and cytogenetic.
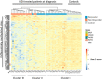
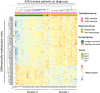
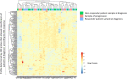
Results: Unsupervised analysis of global methylation pattern at diagnosis did not allow patients to be differentiated according to the cytological subtype, cytogenetic groups, treatment response or patient outcome. However, after a supervised analysis we found a methylation signature defined by 200 probes, which allowed differentiating between patients responding and non-responding to azacitidine (AZA) treatment and a different methylation pattern also defined by 200 probes that allowed to differentiate patients according to their survival. On studying follow-up samples, we confirmed that AZA decreases global DNA methylation, but in our cohort the degree of methylation decrease did not correlate with the type of response. The methylation signature detected at diagnosis was not useful in treated samples to distinguish patients who were going to relapse or progress.
Conclusions: Our findings suggest that in a subset of specific CpGs, altered DNA methylation patterns at diagnosis may be useful as a biomarker for predicting AZA response and survival.
Keywords: Azacitidine; DNA methylation; Epigenetic drugs; Hypomethylating agents; Myelodysplastic syndromes; Prognostic factors; Secondary acute myeloid leukemia.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

