A novel DNA chromatography method to discriminate Mycobacterium abscessus subspecies and macrolide susceptibility
- PMID: 33446475
- PMCID: PMC7910664
- DOI: 10.1016/j.ebiom.2020.103187
A novel DNA chromatography method to discriminate Mycobacterium abscessus subspecies and macrolide susceptibility
Abstract
Background: The clinical impact of infection with Mycobacterium (M.) abscessus complex (MABC), a group of emerging non-tuberculosis mycobacteria (NTM), is increasing. M. abscessus subsp. abscessus/bolletii frequently shows natural resistance to macrolide antibiotics, whereas M. abscessus subsp. massiliense is generally susceptible. Therefore, rapid and accurate discrimination of macrolide-susceptible MABC subgroups is required for effective clinical decisions about macrolide treatments for MABC infection. We aimed to develop a simple and rapid diagnostic that can identify MABC isolates showing macrolide susceptibility.
Methods: Whole genome sequencing (WGS) was performed for 148 clinical or environmental MABC isolates from Japan to identify genetic markers that can discriminate three MABC subspecies and the macrolide-susceptible erm(41) T28C sequevar. Using the identified genetic markers, we established PCR based- or DNA chromatography-based assays. Validation testing was performed using MABC isolates from Taiwan.
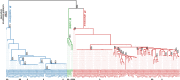
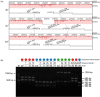
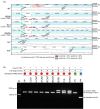
Finding: We identified unique sequence regions that could be used to differentiate the three subspecies. Our WGS-based phylogenetic analysis indicated that M. abscessus carrying the macrolide-susceptible erm(41) T28C sequevar were tightly clustered, and identified 11 genes that were significantly associated with the lineage for use as genetic markers. To detect these genetic markers and the erm(41) locus, we developed a DNA chromatography method that identified three subspecies, the erm(41) T28C sequevar and intact erm(41) for MABC in a single assay within one hour. The agreement rate between the DNA chromatography-based and WGS-based identification was 99·7%.
Interpretation: We developed a novel, rapid and simple DNA chromatography method for identification of MABC macrolide susceptibility with high accuracy.
Funding: AMED, JSPS KAKENHI.
Keywords: ATS/ERS/ESCMID/IDSA guideline; DNA chromatography; Drug susceptibility; Non-tuberculosis mycobacteria; Subspecies discrimination; Whole genome sequencing.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest Mitsunori Yoshida, Sotaro Sano, Shigehiko Miyamoto and Yoshihiko Hoshino are listed on a pending patent in Japan for the DNA chromatography methodology to distinguish MABC and identify macrolide susceptibility (JP2020–066277 and JP2020–066306). Dr. Morimoto and Dr Kurashima report personal fees as Consultant of INSMED, outside the submitted work. The other authors have nothing to disclose under this manuscript.
Figures


References
-
- Oren A., Garrity G.M. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2016;66:2463–2466. - PubMed
-
- Adekambi T., Sassi M., van Ingen J., Drancourt M. Reinstating Mycobacterium massiliense and Mycobacterium bolletii as species of the Mycobacterium abscessus complex. Int J Syst Evol Microbiol. 2017;67:2726–2730. - PubMed
-
- Tortoli E., Kohl T.A., Brown-Elliott B.A., Trovato A., Cardoso-Leão S., Garcia M.J. Mycobacterium abscessus, a taxonomic puzzle. Int J Syst Evol Microbiol. 2018;68:467–469. - PubMed
-
- Griffith D.E., Aksamit T., Brown-Elliott B.A., Catanzaro A., Daley C., Gordin F. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

