Reduced RNA turnover as a driver of cellular senescence
- PMID: 33446491
- PMCID: PMC7812316
- DOI: 10.26508/lsa.202000809
Reduced RNA turnover as a driver of cellular senescence
Abstract
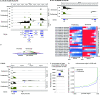
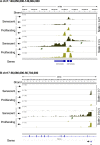
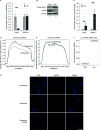
Accumulation of senescent cells is an important contributor to chronic inflammation upon aging. The inflammatory phenotype of senescent cells was previously shown to be driven by cytoplasmic DNA. Here, we propose that cytoplasmic double-stranded RNA has a similar effect. We find that several cell types driven into senescence by different routes share an accumulation of long promoter RNAs and 3' gene extensions rich in retrotransposon sequences. Accordingly, these cells display increased expression of genes involved in response to double stranded RNA of viral origin downstream of the interferon pathway. The RNA accumulation is associated with evidence of reduced RNA turnover, including in some cases, reduced expression of RNA exosome subunits. Reciprocally, depletion of RNA exosome subunit EXOSC3 accelerated expression of multiple senescence markers. A senescence-like RNA accumulation was also observed in cells exposed to oxidative stress, an important trigger of cellular senescence. Altogether, we propose that in a subset of senescent cells, repeat-containing transcripts stabilized by oxidative stress or reduced RNA exosome activity participate in driving and maintaining the permanent inflammatory state characterizing cellular senescence.
© 2021 Mullani et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
