Divergent improvement of two cultivated allotetraploid cotton species
- PMID: 33448110
- PMCID: PMC8313128
- DOI: 10.1111/pbi.13547
Divergent improvement of two cultivated allotetraploid cotton species
Abstract
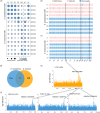
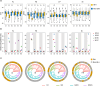
Interspecific genomic variation can provide a genetic basis for local adaptation and domestication. A series of studies have presented its role of interspecific haplotypes and introgressions in adaptive traits, but few studies have addressed their role in improving agronomic character. Two allotetraploid Gossypium species, Gossypium barbadense (Gb) and G. hirsutum (Gh) originating from the Americas, are cultivated independently. Here, through sequencing and the comparison of one GWAS panel in 229 Gb accessions and two GWAS panels in 491 Gh accessions, we found that most associated loci or functional haplotypes for agronomic traits were highly divergent, representing the strong divergent improvement between Gb and Gh. Using a comprehensive interspecific haplotype map, we revealed that six interspecific introgressions from Gh to Gb were significantly associated with the phenotypic performance of Gb, which could explain 5%-40% of phenotypic variation in yield and fibre qualities. In addition, three introgressions overlapped with six associated loci in Gb, indicating that these introgression regions were under further selection and stabilized during improvement. A single interspecific introgression often possessed yield-increasing potential but decreased fibre qualities, or the opposite, making it difficult to simultaneously improve yield and fibre qualities. Our study not only has proved the importance of interspecific functional haplotypes or introgressions in the divergent improvement of Gb and Gh, but also supports their potential value in further human-mediated hybridization or precision breeding.
Keywords: Gossypium species; divergent improvement; interspecific haplotypes; interspecific introgression.
© 2021 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

