Target binding and residence: a new determinant of DNA double-strand break repair pathway choice in CRISPR/Cas9 genome editing
- PMID: 33448189
- PMCID: PMC7818014
- DOI: 10.1631/jzus.B2000282
Target binding and residence: a new determinant of DNA double-strand break repair pathway choice in CRISPR/Cas9 genome editing
Abstract
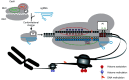
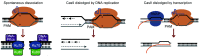
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) is widely used for targeted genomic and epigenomic modifications and imaging in cells and organisms, and holds tremendous promise in clinical applications. The efficiency and accuracy of the technology are partly determined by the target binding affinity and residence time of Cas9-single-guide RNA (sgRNA) at a given site. However, little attention has been paid to the effect of target binding affinity and residence duration on the repair of Cas9-induced DNA double-strand breaks (DSBs). We propose that the choice of DSB repair pathway may be altered by variation in the binding affinity and residence duration of Cas9-sgRNA at the cleaved target, contributing to significantly heterogeneous mutations in CRISPR/Cas9 genome editing. Here, we discuss the effect of Cas9-sgRNA target binding and residence on the choice of DSB repair pathway in CRISPR/Cas9 genome editing, and the opportunity this presents to optimize Cas9-based technology.
Keywords: CRISPR/Cas9 genome editing; Double-strand break (DSB) repair pathway choice; Target binding affinity; Target residence.
Figures
Similar articles
-
Target residence of Cas9-sgRNA influences DNA double-strand break repair pathway choices in CRISPR/Cas9 genome editing.Genome Biol. 2022 Aug 1;23(1):165. doi: 10.1186/s13059-022-02736-5. Genome Biol. 2022. PMID: 35915475 Free PMC article.
-
Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks.Int J Mol Sci. 2020 Sep 4;21(18):6461. doi: 10.3390/ijms21186461. Int J Mol Sci. 2020. PMID: 32899704 Free PMC article. Review.
-
Precision genome editing in the CRISPR era.Biochem Cell Biol. 2017 Apr;95(2):187-201. doi: 10.1139/bcb-2016-0137. Epub 2016 Sep 29. Biochem Cell Biol. 2017. PMID: 28177771 Review.
-
CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations.Nat Commun. 2019 Mar 8;10(1):1136. doi: 10.1038/s41467-019-09006-2. Nat Commun. 2019. PMID: 30850590 Free PMC article.
-
Single-Strand Annealing Plays a Major Role in Double-Strand DNA Break Repair following CRISPR-Cas9 Cleavage in Leishmania.mSphere. 2019 Aug 21;4(4):e00408-19. doi: 10.1128/mSphere.00408-19. mSphere. 2019. PMID: 31434745 Free PMC article.
Cited by
-
Role and application of CRISPR-Cas9 in the management of Alzheimer's disease.Ann Med Surg (Lond). 2024 Jan 5;86(3):1517-1521. doi: 10.1097/MS9.0000000000001692. eCollection 2024 Mar. Ann Med Surg (Lond). 2024. PMID: 38463115 Free PMC article. Review.
-
Catalytically inactive Cas9 attenuates DNA end resection: A potential application for region-restricted random mutagenesis.iScience. 2025 May 20;28(6):112702. doi: 10.1016/j.isci.2025.112702. eCollection 2025 Jun 20. iScience. 2025. PMID: 40520111 Free PMC article.
-
Visualization of a multi-turnover Cas9 after product release.Nat Commun. 2025 Jul 1;16(1):5681. doi: 10.1038/s41467-025-60668-7. Nat Commun. 2025. PMID: 40593576 Free PMC article.
-
A strain of Vibrio alginolyticus isolated from Azumapecten farreri and its pathogenic mechanism using CRISPR-Cas9 technology.Biotechnol Lett. 2023 Oct;45(10):1279-1291. doi: 10.1007/s10529-023-03394-8. Epub 2023 Jul 28. Biotechnol Lett. 2023. PMID: 37505340
-
Target residence of Cas9-sgRNA influences DNA double-strand break repair pathway choices in CRISPR/Cas9 genome editing.Genome Biol. 2022 Aug 1;23(1):165. doi: 10.1186/s13059-022-02736-5. Genome Biol. 2022. PMID: 35915475 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

