A peptide encoded within a 5' untranslated region promotes pain sensitization in mice
- PMID: 33449506
- PMCID: PMC8119312
- DOI: 10.1097/j.pain.0000000000002191
A peptide encoded within a 5' untranslated region promotes pain sensitization in mice
Abstract
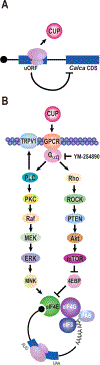
Translational regulation permeates neuronal function. Nociceptors are sensory neurons responsible for the detection of harmful stimuli. Changes in their activity, termed plasticity, are intimately linked to the persistence of pain. Although inhibitors of protein synthesis robustly attenuate pain-associated behavior, the underlying targets that support plasticity are largely unknown. Here, we examine the contribution of protein synthesis in regions of RNA annotated as noncoding. Based on analyses of previously reported ribosome profiling data, we provide evidence for widespread translation in noncoding transcripts and regulatory regions of mRNAs. We identify an increase in ribosome occupancy in the 5' untranslated regions of the calcitonin gene-related peptide (CGRP/Calca). We validate the existence of an upstream open reading frame (uORF) using a series of reporter assays. Fusion of the uORF to a luciferase reporter revealed active translation in dorsal root ganglion neurons after nucleofection. Injection of the peptide corresponding to the calcitonin gene-related peptide-encoded uORF resulted in pain-associated behavioral responses in vivo and nociceptor sensitization in vitro. An inhibitor of heterotrimeric G protein signaling blocks both effects. Collectively, the data suggest pervasive translation in regions of the transcriptome annotated as noncoding in dorsal root ganglion neurons and identify a specific uORF-encoded peptide that promotes pain sensitization through GPCR signaling.
Copyright © 2021 International Association for the Study of Pain.
Conflict of interest statement
Declaration interests
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

