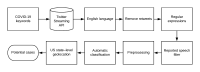
Toward Using Twitter for Tracking COVID-19: A Natural Language Processing Pipeline and Exploratory Data Set
- PMID: 33449904
- PMCID: PMC7834613
- DOI: 10.2196/25314
Toward Using Twitter for Tracking COVID-19: A Natural Language Processing Pipeline and Exploratory Data Set
Abstract
Background: In the United States, the rapidly evolving COVID-19 outbreak, the shortage of available testing, and the delay of test results present challenges for actively monitoring its spread based on testing alone.
Objective: The objective of this study was to develop, evaluate, and deploy an automatic natural language processing pipeline to collect user-generated Twitter data as a complementary resource for identifying potential cases of COVID-19 in the United States that are not based on testing and, thus, may not have been reported to the Centers for Disease Control and Prevention.
Methods: Beginning January 23, 2020, we collected English tweets from the Twitter Streaming application programming interface that mention keywords related to COVID-19. We applied handwritten regular expressions to identify tweets indicating that the user potentially has been exposed to COVID-19. We automatically filtered out "reported speech" (eg, quotations, news headlines) from the tweets that matched the regular expressions, and two annotators annotated a random sample of 8976 tweets that are geo-tagged or have profile location metadata, distinguishing tweets that self-report potential cases of COVID-19 from those that do not. We used the annotated tweets to train and evaluate deep neural network classifiers based on bidirectional encoder representations from transformers (BERT). Finally, we deployed the automatic pipeline on more than 85 million unlabeled tweets that were continuously collected between March 1 and August 21, 2020.
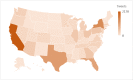
Results: Interannotator agreement, based on dual annotations for 3644 (41%) of the 8976 tweets, was 0.77 (Cohen κ). A deep neural network classifier, based on a BERT model that was pretrained on tweets related to COVID-19, achieved an F1-score of 0.76 (precision=0.76, recall=0.76) for detecting tweets that self-report potential cases of COVID-19. Upon deploying our automatic pipeline, we identified 13,714 tweets that self-report potential cases of COVID-19 and have US state-level geolocations.
Conclusions: We have made the 13,714 tweets identified in this study, along with each tweet's time stamp and US state-level geolocation, publicly available to download. This data set presents the opportunity for future work to assess the utility of Twitter data as a complementary resource for tracking the spread of COVID-19.
Keywords: COVID-19; coronavirus; data mining; epidemiology; infodemiology; natural language processing; pandemics; social media.
©Ari Z Klein, Arjun Magge, Karen O'Connor, Jesus Ivan Flores Amaro, Davy Weissenbacher, Graciela Gonzalez Hernandez. Originally published in the Journal of Medical Internet Research (http://www.jmir.org), 22.01.2021.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
Similar articles
-
A natural language processing pipeline to advance the use of Twitter data for digital epidemiology of adverse pregnancy outcomes.J Biomed Inform. 2020;112S:100076. doi: 10.1016/j.yjbinx.2020.100076. Epub 2020 Aug 8. J Biomed Inform. 2020. PMID: 34417007 Free PMC article.
-
Automatically Identifying Twitter Users for Interventions to Support Dementia Family Caregivers: Annotated Data Set and Benchmark Classification Models.JMIR Aging. 2022 Sep 16;5(3):e39547. doi: 10.2196/39547. JMIR Aging. 2022. PMID: 36112408 Free PMC article.
-
Developing an Automatic System for Classifying Chatter About Health Services on Twitter: Case Study for Medicaid.J Med Internet Res. 2021 May 3;23(5):e26616. doi: 10.2196/26616. J Med Internet Res. 2021. PMID: 33938807 Free PMC article.
-
Toward Using Twitter for PrEP-Related Interventions: An Automated Natural Language Processing Pipeline for Identifying Gay or Bisexual Men in the United States.JMIR Public Health Surveill. 2022 Apr 25;8(4):e32405. doi: 10.2196/32405. JMIR Public Health Surveill. 2022. PMID: 35468092 Free PMC article.
-
Methods to Establish Race or Ethnicity of Twitter Users: Scoping Review.J Med Internet Res. 2022 Apr 29;24(4):e35788. doi: 10.2196/35788. J Med Internet Res. 2022. PMID: 35486433 Free PMC article.
Cited by
-
A chronological and geographical analysis of personal reports of COVID-19 on Twitter from the UK.Digit Health. 2022 May 5;8:20552076221097508. doi: 10.1177/20552076221097508. eCollection 2022 Jan-Dec. Digit Health. 2022. PMID: 35574580 Free PMC article.
-
Forecasting the COVID-19 Epidemic by Integrating Symptom Search Behavior Into Predictive Models: Infoveillance Study.J Med Internet Res. 2021 Aug 11;23(8):e28876. doi: 10.2196/28876. J Med Internet Res. 2021. PMID: 34156966 Free PMC article.
-
Perspectives of the COVID-19 Pandemic on Reddit: Comparative Natural Language Processing Study of the United States, the United Kingdom, Canada, and Australia.JMIR Infodemiology. 2022 Sep 27;2(2):e36941. doi: 10.2196/36941. eCollection 2022 Jul-Dec. JMIR Infodemiology. 2022. PMID: 36196144 Free PMC article.
-
Analysis of social media data for public emotion on the Wuhan lockdown event during the COVID-19 pandemic.Comput Methods Programs Biomed. 2021 Nov;212:106468. doi: 10.1016/j.cmpb.2021.106468. Epub 2021 Oct 14. Comput Methods Programs Biomed. 2021. PMID: 34715513 Free PMC article.
-
Implicit Incentives Among Reddit Users to Prioritize Attention Over Privacy and Reveal Their Faces When Discussing Direct-to-Consumer Genetic Test Results: Topic and Attention Analysis.JMIR Infodemiology. 2022 Aug 3;2(2):e35702. doi: 10.2196/35702. eCollection 2022 Jul-Dec. JMIR Infodemiology. 2022. PMID: 37113452 Free PMC article.
References
-
- Menni C, Valdes AM, Freidin MB, Sudre CH, Nguyen LH, Drew DA, Ganesh S, Varsavsky T, Cardoso MJ, El-Sayed Moustafa JS, Visconti A, Hysi P, Bowyer RCE, Mangino M, Falchi M, Wolf J, Ourselin S, Chan AT, Steves CJ, Spector TD. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med. 2020 Jul 11;26(7):1037–1040. doi: 10.1038/s41591-020-0916-2. http://europepmc.org/abstract/MED/32393804 - DOI - PMC - PubMed
-
- Smith A, Anderson M. Social media use in 2018. Pew Research Center. 2018. Mar 01, [2020-09-29]. https://www.pewresearch.org/internet/2018/03/01/social-media-use-in-2018/
-
- Jeon J, Baruah G, Sarabadani S, Palanica A. Identification of Risk Factors and Symptoms of COVID-19: Analysis of Biomedical Literature and Social Media Data. J Med Internet Res. 2020 Oct 02;22(10):e20509. doi: 10.2196/20509. https://www.jmir.org/2020/10/e20509/ - DOI - PMC - PubMed
-
- Mackey T, Purushothaman V, Li J, Shah N, Nali M, Bardier C, Liang B, Cai M, Cuomo R. Machine Learning to Detect Self-Reporting of Symptoms, Testing Access, and Recovery Associated With COVID-19 on Twitter: Retrospective Big Data Infoveillance Study. JMIR Public Health Surveill. 2020 Jun 08;6(2):e19509. doi: 10.2196/19509. https://publichealth.jmir.org/2020/2/e19509/ - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical