Functional screening of a Caatinga goat (Capra hircus) rumen metagenomic library reveals a novel GH3 β-xylosidase
- PMID: 33449963
- PMCID: PMC7810302
- DOI: 10.1371/journal.pone.0245118
Functional screening of a Caatinga goat (Capra hircus) rumen metagenomic library reveals a novel GH3 β-xylosidase
Abstract
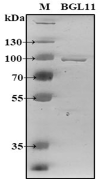
Functional screening of metagenomic libraries is an effective approach for identification of novel enzymes. A Caatinga biome goat rumen metagenomic library was screened using esculin as a substrate, and a gene from an unknown bacterium encoding a novel GH3 enzyme, BGL11, was identified. None of the BGL11 closely related genes have been previously characterized. Recombinant BGL11 was obtained and kinetically characterized. Substrate specificity of the purified protein was assessed using seven synthetic aryl substrates. Activity towards nitrophenyl-β-D-glucopyranoside (pNPG), 4-nitrophenyl-β-D-xylopyranoside (pNPX) and 4-nitrophenyl-β-D-cellobioside (pNPC) suggested that BGL11 is a multifunctional enzyme with β-glucosidase, β-xylosidase, and cellobiohydrolase activities. However, further testing with five natural substrates revealed that, although BGL11 has multiple substrate specificity, it is most active towards xylobiose. Thus, in its native goat rumen environment, BGL11 most likely functions as an extracellular β-xylosidase acting on hemicellulose. Biochemical characterization of BGL11 showed an optimal pH of 5.6, and an optimal temperature of 50°C. Enzyme stability, an important parameter for industrial application, was also investigated. At 40°C purified BGL11 remained active for more than 15 hours without reduction in activity, and at 50°C, after 7 hours of incubation, BGL11 remained 60% active. The enzyme kinetic parameters of Km and Vmax using xylobiose were determined to be 3.88 mM and 38.53 μmol.min-1.mg-1, respectively, and the Kcat was 57.79 s-1. In contrast to BLG11, most β-xylosidases kinetically studied belong to the GH43 family and have been characterized only using synthetic substrates. In industry, β-xylosidases can be used for plant biomass deconstruction, and the released sugars can be fermented into valuable bio-products, ranging from the biofuel ethanol to the sugar substitute xylitol.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Zhang S, Wang H, Shi P, Xu B, Bai Y, Luo H, et al. Cloning, expression, and characterization of a thermostable β-xylosidase from thermoacidophilic Alicyclobacillus sp. A4. Process Biochem. 2014; 49: 1422–1428. 10.1016/j.procbio.2014.05.020 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

