Evidence for multi-copy Mega-NUMTs in the human genome
- PMID: 33450006
- PMCID: PMC7897518
- DOI: 10.1093/nar/gkaa1271
Evidence for multi-copy Mega-NUMTs in the human genome
Abstract
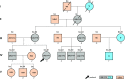
The maternal mode of mitochondrial DNA (mtDNA) inheritance is central to human genetics. Recently, evidence for bi-parental inheritance of mtDNA was claimed for individuals of three pedigrees that suffered mitochondrial disorders. We sequenced mtDNA using both direct Sanger and Massively Parallel Sequencing in several tissues of eleven maternally related and other affiliated healthy individuals of a family pedigree and observed mixed mitotypes in eight individuals. Cells without nuclear DNA, i.e. thrombocytes and hair shafts, only showed the mitotype of haplogroup (hg) V. Skin biopsies were prepared to generate ρ° cells void of mtDNA, sequencing of which resulted in a hg U4c1 mitotype. The position of the Mega-NUMT sequence was determined by fluorescence in situ hybridization and two different quantitative PCR assays were used to determine the number of contributing mtDNA copies. Thus, evidence for the presence of repetitive, full mitogenome Mega-NUMTs matching haplogroup U4c1 in various tissues of eight maternally related individuals was provided. Multi-copy Mega-NUMTs mimic mixtures of mtDNA that cannot be experimentally avoided and thus may appear in diverse fields of mtDNA research and diagnostics. We demonstrate that hair shaft mtDNA sequencing provides a simple but reliable approach to exclude NUMTs as source of misleading results.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Hutchison C.A., Newbold J.B., Potter S.S., Edgell M.H.. Maternal inheritance of mammalian mitochondrial DNA. Nature. 1974; 251:536–538. - PubMed
-
- Passamonti M., Ghiselli F.. Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 2009; 28:79–89. - PubMed
-
- Cann R.L., Stoneking M., Wilson A.C.. Mitochondrial DNA and human evolution. Nature. 1987; 325:31–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

