The Molecular Determinants of Thermoadaptation: Methanococcales as a Case Study
- PMID: 33450027
- PMCID: PMC8097290
- DOI: 10.1093/molbev/msaa312
The Molecular Determinants of Thermoadaptation: Methanococcales as a Case Study
Abstract
Previous reports have shown that environmental temperature impacts proteome evolution in Bacteria and Archaea. However, it is unknown whether thermoadaptation mainly occurs via the sequential accumulation of substitutions, massive horizontal gene transfers, or both. Measuring the real contribution of amino acid substitution to thermoadaptation is challenging, because of confounding environmental and genetic factors (e.g., pH, salinity, genomic G + C content) that also affect proteome evolution. Here, using Methanococcales, a major archaeal lineage, as a study model, we show that optimal growth temperature is the major factor affecting variations in amino acid frequencies of proteomes. By combining phylogenomic and ancestral sequence reconstruction approaches, we disclose a sequential substitutional scheme in which lysine plays a central role by fine tuning the pool of arginine, serine, threonine, glutamine, and asparagine, whose frequencies are strongly correlated with optimal growth temperature. Finally, we show that colonization to new thermal niches is not associated with high amounts of horizontal gene transfers. Altogether, although the acquisition of a few key proteins through horizontal gene transfer may have favored thermoadaptation in Methanococcales, our findings support sequential amino acid substitutions as the main factor driving thermoadaptation.
Keywords: Methanococci; ancestral sequence reconstruction; evolutionary rates; extremophiles; horizontal gene transfer; prokaryotes; protein.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

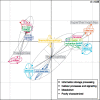

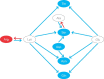

References
-
- Albers SV, Driessen AJ.. 2007. Membrane adaptations of (hyper)thermophiles to high temperatures. In: Gerday C, Glansdorff N, editors. Physiology and biochemistry of extremophiles Washington, D.C.: ASM press. p. 104–116.
-
- Albers S-V, Vossenberg JL, Driessen AJ, Konings WN.. 2001. Bioenergetics and solute uptake under extreme conditions. Extremophiles 5(5):285–294. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

