Sequence analysis of Indian SARS-CoV-2 isolates shows a stronger interaction of mutant receptor-binding domain with ACE2
- PMID: 33450373
- PMCID: PMC7833473
- DOI: 10.1016/j.ijid.2021.01.020
Sequence analysis of Indian SARS-CoV-2 isolates shows a stronger interaction of mutant receptor-binding domain with ACE2
Abstract
Objective: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected the whole world, including Odisha, a state in eastern India. Many people have migrated to the state from different countries as well as other states during this SARS-CoV-2 pandemic. The aim of this study was to analyse the receptor-binding domain (RBD) sequence of the spike protein from isolates collected from throat swab samples of COVID-19-positive patients and further to assess the RBD affinity for angiotensin-converting enzyme 2 (ACE2) of different species, including humans.
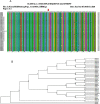
Methods: Whole-genome sequencing for 35 clinical SARS-CoV-2 isolates from COVID-19-positive patients was performed by ARTIC amplicon-based sequencing. Sequence analysis and phylogenetic analysis were performed for the spike region and the RBD region of all isolates. The interaction between the RBD and ACE2 of five different species was also analysed.
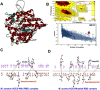
Results: The spike region of 32 isolates showed one or multiple alterations in nucleotide bases in comparison with the Wuhan reference strain. One of the identified mutations, at position 1204 (Ref A, RMRC 22 C), in the RBD coding region of the spike protein showed stronger binding affinity for human ACE2. Furthermore, RBDs of all the Indian isolates showed binding affinity for ACE2 of different species.
Conclusion: As mutant RBD showed stronger interaction with human ACE2, it could potentially result in higher infectivity. The binding affinity of the RBDs for ACE2 of all five species studied suggests that the virus can infect a wide variety of animals, which could also act as natural reservoir for SARS-CoV-2.
Keywords: ACE2; Indian isolates; Receptor-binding domain; SARS-CoV-2; Spike.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures




References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Benkert P., Tosatto S.C.E., Schomburg D. QMEAN: a comprehensive scoring function for model quality assessment. Protein Struct Funct Genet. 2008;71:261–277. - PubMed
-
- Bowie J.U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

