Overcoming Supply Shortage for SARS-CoV-2 Detection by RT-qPCR
- PMID: 33450867
- PMCID: PMC7828326
- DOI: 10.3390/genes12010090
Overcoming Supply Shortage for SARS-CoV-2 Detection by RT-qPCR
Abstract
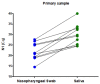
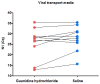
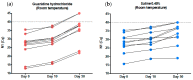
In February 2020, our laboratory started to offer a RT-qPCR assay for the qualitative detection of severe acute respiratory syndrome coronavirus 2. A few months after the assay was released to our patients, some materials, reagents, and equipment became in short supply. Alternative protocols were necessary in order to avoid stopping testing to the population. However, the suitability of these alternatives needs to be validated before their use. Here, we investigated if saliva is a reliable alternative specimen to nasopharyngeal swabs; if 0.45% saline is a reliable alternative to guanidine hydrochloride as a collection viral transport media; the stability of SARS-COV-2 in guanidine hydrochloride and in 0.45% saline for 10 and 50 days at room temperature; and if the primers/probe concentration and thermocycling times could be reduced so as to overcome the short supply of these reagents and equipment, without a significant loss of the assay performance. We found that saliva is not an appropriated specimen for our method-nasopharyngeal swabs perform better. Saline (0.45%) and guanidine hydrochloride have a similar SARS-CoV-2 diagnostic capability as tube additives. Reliable SARS-CoV-2 RNA detection can be performed after sample storage for 10 days at room temperature (18-23 °C) in both 0.45% saline and guanidine hydrochloride. Using synthetic RNA, and decreasing the concentration of primers by five-fold and probes by 2.5-fold, changed the assay limit of detection (LOD) from 7.2 copies/reaction to 23.7 copies/reaction and the subsequent reducing of thermocycling times changed the assay LOD from 23.7 copies/reaction to 44.2 copies/reaction. However, using real clinical samples with Cq values ranging from ~12.15 to ~36.46, the results of the three tested conditions were almost identical. These alterations will not affect the vast majority of diagnostics and increase the daily testing capability in 30% and increase primers and probe stocks in 500% and 250%, respectively. Taken together, the alternative protocols described here overcome the short supply of tubes, reagents and equipment during the SARS-CoV-2 pandemic, avoiding the collapse of test offering for the population: 105,757 samples were processed, and 25,156 SARS-CoV-2 diagnostics were performed from 9 May 2020 to 30 June 2020.
Keywords: RT-qPCR; SARS-CoV-2; validation.
Conflict of interest statement
Sabin laboratory is a private commercial laboratory. All authors are employees of Sabin Laboratory. The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

