The Unique, the Known, and the Unknown of Spumaretrovirus Assembly
- PMID: 33451128
- PMCID: PMC7828637
- DOI: 10.3390/v13010105
The Unique, the Known, and the Unknown of Spumaretrovirus Assembly
Abstract
Within the family of Retroviridae, foamy viruses (FVs) are unique and unconventional with respect to many aspects in their molecular biology, including assembly and release of enveloped viral particles. Both components of the minimal assembly and release machinery, Gag and Env, display significant differences in their molecular structures and functions compared to the other retroviruses. This led to the placement of FVs into a separate subfamily, the Spumaretrovirinae. Here, we describe the molecular differences in FV Gag and Env, as well as Pol, which is translated as a separate protein and not in an orthoretroviral manner as a Gag-Pol fusion protein. This feature further complicates FV assembly since a specialized Pol encapsidation strategy via a tripartite Gag-genome-Pol complex is used. We try to relate the different features and specific interaction patterns of the FV Gag, Pol, and Env proteins in order to develop a comprehensive and dynamic picture of particle assembly and release, but also other features that are indirectly affected. Since FVs are at the root of the retrovirus tree, we aim at dissecting the unique/specialized features from those shared among the Spuma- and Orthoretrovirinae. Such analyses may shed light on the evolution and characteristics of virus envelopment since related viruses within the Ortervirales, for instance LTR retrotransposons, are characterized by different levels of envelopment, thus affecting the capacity for intercellular transmission.
Keywords: Env leader protein; Ortervirales; Pol protein packaging; RNA-mediated Pol tethering; assembly; foamy virus; particle budding; retrovirus evolution; spumavirus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
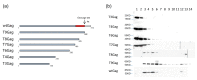




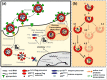
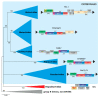
References
-
- Goff S.P. Retroviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. 6th ed. Volume 2. Lippincott Williams & Wilkins, a Wolters Kluwer Business; Philadelphia, PA, USA: 2013. pp. 1424–1473.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

