Gene expression feature selection for prostate cancer diagnosis using a two-phase heuristic-deterministic search strategy
- PMID: 33451186
- PMCID: PMC8687315
- DOI: 10.1049/iet-syb.2017.0044
Gene expression feature selection for prostate cancer diagnosis using a two-phase heuristic-deterministic search strategy
Abstract
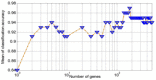
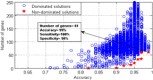
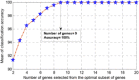
Here, a two-phase search strategy is proposed to identify the biomarkers in gene expression data set for the prostate cancer diagnosis. A statistical filtering method is initially employed to remove the noisiest data. In the first phase of the search strategy, a multi-objective optimisation based on the binary particle swarm optimisation algorithm tuned by a chaotic method is proposed to select the optimal subset of genes with the minimum number of genes and the maximum classification accuracy. Finally, in the second phase of the search strategy, the cache-based modification of the sequential forward floating selection algorithm is used to find the most discriminant genes from the optimal subset of genes selected in the first phase. The results of applying the proposed algorithm on the available challenging prostate cancer data set demonstrate that the proposed algorithm can perfectly identify the informative genes such that the classification accuracy, sensitivity, and specificity of 100% are achieved with only nine biomarkers.
Keywords: available challenging prostate cancer data; biological organs; biomarkers; cancer; chaotic method; discriminant genes; feature extraction; gene expression data; gene expression feature selection; genetics; heuristic-deterministic search strategy; informative genes; multiobjective optimisation; noisiest data; optimisation; particle swarm optimisation; particle swarm optimisation algorithm; pattern classification; prostate cancer diagnosis; search problems; selection algorithm; statistical filtering method; two-phase search strategy.
© 2020 The Institution of Engineering and Technology.
Figures

References
-
- Lv J., Peng Q., and Chen X. et al.: ‘A multi‐objective heuristic algorithm for gene expression microarray data classification’, Expert Syst. Appl., 2016, 59, pp. 13–19
-
- Sharma A., Imoto S., and Miyano S.: ‘A top‐R feature selection algorithm for microarray gene expression data’, IEEE/ACM Trans. Comput. Biol. Bioinform. (TCBB), 2012, 9, (3), pp. 754–764 - PubMed
-
- Nguyen T., Khosravi A., and Creighton D. et al.: ‘Hidden Markov models for cancer classification using gene expression profiles’, Inf. Sci., 2015, 316, pp. 293–307
-
- Wang L., and Han B.: ‘Hybrid feature selection method for gene expression analysis’, Electron. Lett., 2014, 50, (18), pp. 1269–1271
-
- Cui Y., Zheng C.‐H., and Yang J. et al.: ‘Sparse maximum margin discriminant analysis for feature extraction and gene selection on gene expression data’, Comput. Biol. Med., 2013, 43, (7), pp. 933–941 - PubMed
LinkOut - more resources
Full Text Sources

