NIST Interlaboratory Study on Glycosylation Analysis of Monoclonal Antibodies: Comparison of Results from Diverse Analytical Methods
- PMID: 31591262
- PMCID: PMC6944243
- DOI: 10.1074/mcp.RA119.001677
NIST Interlaboratory Study on Glycosylation Analysis of Monoclonal Antibodies: Comparison of Results from Diverse Analytical Methods
Abstract
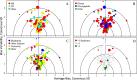
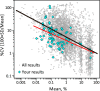
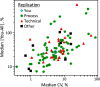
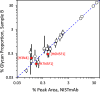
Glycosylation is a topic of intense current interest in the development of biopharmaceuticals because it is related to drug safety and efficacy. This work describes results of an interlaboratory study on the glycosylation of the Primary Sample (PS) of NISTmAb, a monoclonal antibody reference material. Seventy-six laboratories from industry, university, research, government, and hospital sectors in Europe, North America, Asia, and Australia submitted a total of 103 reports on glycan distributions. The principal objective of this study was to report and compare results for the full range of analytical methods presently used in the glycosylation analysis of mAbs. Therefore, participation was unrestricted, with laboratories choosing their own measurement techniques. Protein glycosylation was determined in various ways, including at the level of intact mAb, protein fragments, glycopeptides, or released glycans, using a wide variety of methods for derivatization, separation, identification, and quantification. Consequently, the diversity of results was enormous, with the number of glycan compositions identified by each laboratory ranging from 4 to 48. In total, one hundred sixteen glycan compositions were reported, of which 57 compositions could be assigned consensus abundance values. These consensus medians provide community-derived values for NISTmAb PS. Agreement with the consensus medians did not depend on the specific method or laboratory type. The study provides a view of the current state-of-the-art for biologic glycosylation measurement and suggests a clear need for harmonization of glycosylation analysis methods.
Keywords: Glycomics; NISTmAb; fluorescence; glycan; glycopeptide; glycoproteins; glycosylation; interlaboratory study; mass spectrometry; reference antibody.
© 2020 De Leoz et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article. Any mention of commercial products is for information only; it does not imply recommendation or endorsement by any of the participating institutions. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the participating institutions
Figures





References
-
- Dotz V., Haselberg R., Shubhakar A., Kozak R. P., Falck D., Rombouts Y., Reusch D., Somsen G. W., Fernandes D. L., and Wuhrer M. (2015) Mass spectrometry for glycosylation analysis of biopharmaceuticals. Trac-Trend Anal. Chem. 73, 1–9
-
- O'Flaherty R., Muniyappa M., Walsh I., Stockmann H., Hutson R., Saldova R., and Rudd P. M. (2016) High-throughput sequential glycoprofiling of six abundant glycoproteins IgG, IgA, IgM, transferrin, haptoglobin and alpha-1-antitrypsin in ovarian cancer. Glycobiology 26, 1430–1431

