Fecal Microbiota Nutrient Utilization Potential Suggests Mucins as Drivers for Initial Gut Colonization of Mother-Child-Shared Bacteria
- PMID: 33452029
- PMCID: PMC8105027
- DOI: 10.1128/AEM.02201-20
Fecal Microbiota Nutrient Utilization Potential Suggests Mucins as Drivers for Initial Gut Colonization of Mother-Child-Shared Bacteria
Abstract
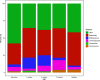
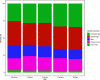
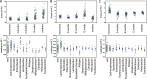
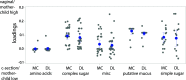
The nutritional drivers for mother-child sharing of bacteria and the corresponding longitudinal trajectory of the infant gut microbiota development are not yet completely settled. We therefore aimed to characterize the mother-child sharing and the inferred nutritional utilization potential for the gut microbiota from a large unselected cohort. We analyzed in depth gut microbiota in 100 mother-child pairs enrolled antenatally from the general population-based Preventing Atopic Dermatitis and Allergies in Children (PreventADALL) cohort. Fecal samples collected at gestational week 18 for mothers and at birth (meconium), 3, 6, and 12 months for infants were analyzed by reduced metagenome sequencing to determine metagenome size and taxonomic composition. The nutrient utilization potential was determined based on the Virtual Metabolic Human (VMH, www.vmh.life) database. The estimated median metagenome size was ∼150 million base pairs (bp) for mothers and ∼20 million bp at birth for the children. Longitudinal analyses revealed mother-child sharing (P < 0.05, chi-square test) from birth up to 6 months for 3 prevalent Bacteroides species (prevalence, >25% for all age groups). In a multivariate analysis of variance (ANOVA), the mother-child-shared Bacteroides were associated with vaginal delivery (1.7% explained variance, P = 0.0001). Both vaginal delivery and mother-child sharing were associated with host-derived mucins as nutrient sources. The age-related increase in metagenome size corresponded to an increased diversity in nutrient utilization, with dietary polysaccharides as the main age-related factor. Our results support host-derived mucins as potential selection means for mother-child sharing of initial colonizers, while the age-related increase in diversity was associated with dietary polysaccharides.IMPORTANCE The initial bacterial colonization of human infants is crucial for lifelong health. Understanding the factors driving this colonization will therefore be of great importance. Here, we used a novel high-taxonomic-resolution approach to deduce the nutrient utilization potential of the infant gut microbiota in a large longitudinal mother-child cohort. We found mucins as potential selection means for the initial colonization of mother-child-shared bacteria, while the transition to a more adult-like microbiota was associated with dietary polysaccharide utilization potential. This knowledge will be important for a future understanding of the importance of diet in shaping the gut microbiota composition and development during infancy.
Keywords: infant gut microbiota.
Copyright © 2021 American Society for Microbiology.
Figures




References
-
- Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, Selvenius J, Oikarinen S, Hyoty H, Virtanen SM, Ilonen J, Ferretti P, Pasolli E, Tett A, Asnicar F, Segata N, Vlamakis H, Lander ES, Huttenhower C, Knip M, Xavier RJ. 2018. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 24:146–154.e4. doi: 10.1016/j.chom.2018.06.007. - DOI - PMC - PubMed
-
- Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, Muzny D, Gibbs RA, Vatanen T, Huttenhower C, Xavier RJ, Rewers M, Hagopian W, Toppari J, Ziegler A-G, She J-X, Akolkar B, Lernmark A, Hyoty H, Vehik K, Krischer JP, Petrosino JF. 2018. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562:583–588. doi: 10.1038/s41586-018-0617-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

