Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS-CoV-2 infection
- PMID: 33452205
- PMCID: PMC7865145
- DOI: 10.1073/pnas.2021579118
Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS-CoV-2 infection
Abstract
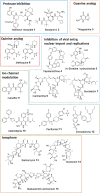
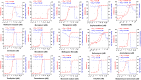
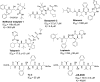
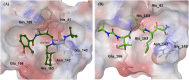
The outbreak of COVID-19 caused by SARS-CoV-2 has resulted in more than 50 million confirmed cases and over 1 million deaths worldwide as of November 2020. Currently, there are no effective antivirals approved by the Food and Drug Administration to contain this pandemic except the antiviral agent remdesivir. In addition, the trimeric spike protein on the viral surface is highly glycosylated and almost 200,000 variants with mutations at more than 1,000 positions in its 1,273 amino acid sequence were reported, posing a major challenge in the development of antibodies and vaccines. It is therefore urgently needed to have alternative and timely treatments for the disease. In this study, we used a cell-based infection assay to screen more than 3,000 agents used in humans and animals, including 2,855 small molecules and 190 traditional herbal medicines, and identified 15 active small molecules in concentrations ranging from 0.1 nM to 50 μM. Two enzymatic assays, along with molecular modeling, were then developed to confirm those targeting the virus 3CL protease and the RNA-dependent RNA polymerase. Several water extracts of herbal medicines were active in the cell-based assay and could be further developed as plant-derived anti-SARS-CoV-2 agents. Some of the active compounds identified in the screen were further tested in vivo, and it was found that mefloquine, nelfinavir, and extracts of Ganoderma lucidum (RF3), Perilla frutescens, and Mentha haplocalyx were effective in a challenge study using hamsters as disease model.
Keywords: SARS-CoV-2; antiviral; cell-based and animal studies; drug repurposing.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
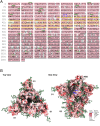
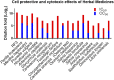

References
-
- Del Rio C., Malani P. N., COVID-19—New insights on a rapidly changing epidemic. JAMA 323, 1339–1340 (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

