Novel dichloromethane-fermenting bacteria in the Peptococcaceae family
- PMID: 33452483
- PMCID: PMC8163858
- DOI: 10.1038/s41396-020-00881-y
Novel dichloromethane-fermenting bacteria in the Peptococcaceae family
Abstract
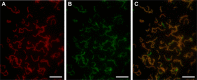
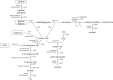
Dichloromethane (DCM; CH2Cl2) is a toxic groundwater pollutant that also has a detrimental effect on atmospheric ozone levels. As a dense non-aqueous phase liquid, DCM migrates vertically through groundwater to low redox zones, yet information on anaerobic microbial DCM transformation remains scarce due to a lack of cultured organisms. We report here the characterisation of DCMF, the dominant organism in an anaerobic enrichment culture (DFE) capable of fermenting DCM to the environmentally benign product acetate. Stable carbon isotope experiments demonstrated that the organism assimilated carbon from DCM and bicarbonate via the Wood-Ljungdahl pathway. DCMF is the first anaerobic DCM-degrading population also shown to metabolise non-chlorinated substrates. It appears to be a methylotroph utilising the Wood-Ljungdahl pathway for metabolism of methyl groups from methanol, choline, and glycine betaine. The flux of these substrates from subsurface environments may either directly (DCM, methanol) or indirectly (choline, glycine betaine) affect the climate. Community profiling and cultivation of cohabiting taxa in culture DFE without DCMF suggest that DCMF is the sole organism in this culture responsible for substrate metabolism, while the cohabitants persist via necromass recycling. Genomic and physiological evidence support placement of DCMF in a novel genus within the Peptococcaceae family, 'Candidatus Formimonas warabiya'.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Shestakova M, Sillanpää M. Removal of dichloromethane from ground and wastewater: a review. Chemosphere. 2013;93:1258–67. - PubMed
-
- Gribble GW. Naturally occurring organohalogen compounds: a comprehensive update. In: Progress in the Chemistry of Organic Natural Products. Volume 91. Vienna, Austria: Springer Vienna; 2010. p. 12–3.
-
- Carpenter LJ, Reimann S. Update on Ozone-Depleting Substances (ODSs) and Other Gases of Interest to the Montreal Protocol. In: Engel A, Montzka SA, editors. Scientific Assessment of Ozone Depletion: 2014. Global Ozone Research and Monitoring Project - Report No 55. Geneva, Switzerland: World Meteotological Organization; 2014.
-
- Hossaini R, Chipperfield MP, Montzka SA, Rap A, Dhomse S, Feng W. Efficiency of short-lived halogens at influencing climate through depletion of stratospheric ozone. Nat Geosci. 2015;8:186–90.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

