Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment
- PMID: 33453151
- PMCID: PMC8026516
- DOI: 10.1016/j.stem.2020.12.004
Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment
Abstract
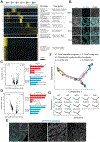
Human testis development in prenatal life involves complex changes in germline and somatic cell identity. To better understand, we profiled and analyzed ∼32,500 single-cell transcriptomes of testicular cells from embryonic, fetal, and infant stages. Our data show that at 6-7 weeks postfertilization, as the testicular cords are established, the Sertoli and interstitial cells originate from a common heterogeneous progenitor pool, which then resolves into fetal Sertoli cells (expressing tube-forming genes) or interstitial cells (including Leydig-lineage cells expressing steroidogenesis genes). Almost 10 weeks later, beginning at 14-16 weeks postfertilization, the male primordial germ cells exit mitosis, downregulate pluripotent transcription factors, and transition into cells that strongly resemble the state 0 spermatogonia originally defined in the infant and adult testes. Therefore, we called these fetal spermatogonia "state f0." Overall, we reveal multiple insights into the coordinated and temporal development of the embryonic, fetal, and postnatal male germline together with the somatic niche.
Keywords: Leydig cell; Sertoli cell; fetal testis development; interstitial cell; primordial germ cell; single-cell RNA sequencing; spermatogonial stem cell.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

