A lowly populated, transient β-sheet structure in monomeric Aβ1-42 identified by multinuclear NMR of chemical denaturation
- PMID: 33453683
- PMCID: PMC7878406
- DOI: 10.1016/j.bpc.2020.106531
A lowly populated, transient β-sheet structure in monomeric Aβ1-42 identified by multinuclear NMR of chemical denaturation
Abstract
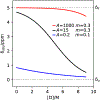
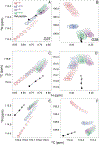
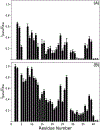
Chemical denaturation is a well-established approach for probing the equilibrium between folded and unfolded states of proteins. We demonstrate applicability of this method to the detection of a small population of a transiently folded structural element in a system that is often considered to be intrinsically fully disordered. The 1HN, 15N, 13Cα, and 13C' chemical shifts of Aβ1-40 and Aβ1-42 peptides and their M35-oxidized variants were monitored as a function of urea concentration and compared to analogous urea titrations of synthetic pentapeptides of homologous sequence. Fitting of the chemical shift titrations yields a 10 ± 1% population for a structured element at the C-terminus of Aβ1-42 that folds with a cooperativity of m = 0.06 kcal/mol·M. The fit also yields the chemical shifts of the folded state and, using a database search, for Aβ1-42 these shifts identified an antiparallel intramolecular β-sheet for residues I32-A42, linked by a type I' β-turn at G37 and G38. The structure is destabilized by oxidation of M35. Paramagnetic relaxation rates and two previously reported weak, medium-range NOE interactions are consistent with this transient β-sheet. Introduction of the requisite A42C mutation and tagging with MTSL resulted in a small stabilization of this β-sheet. Chemical shift analysis suggests a C-terminal β-sheet may be present in Aβ1-40 too, but the turn type at G37 is not type I'. The approach to derive Transient Structure from chemical Denaturation by NMR (TSD-NMR), demonstrated here for Aβ peptides, provides a sensitive tool for identifying the presence of lowly populated, transiently ordered elements in proteins that are considered to be intrinsically disordered, and permits extraction of structural data for such elements.
Keywords: Chemical shift perturbation; Intrinsically disordered protein; Paramagnetic relaxation enhancement; Protein folding; Triple resonance NMR; Urea denaturation.
Published by Elsevier B.V.
Figures



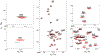

References
-
- Meisl G, Yang X, Hellstrand E, Frohm B, Kirkegaard JB, Cohen SIA, Dobson CM, Linse S, and Knowles TPJ, Differences in nucleation behavior underlie the contrasting aggregation kinetics of the A beta 40 and A beta 42 peptides. Proc. Natl. Acad. Sci. U. S. A, 2014. 111(26): p. 9384–9389. - PMC - PubMed
-
- van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, Pappu RV, Tompa P, Uversky VN, Wright PE, and Babu MM, Classification of Intrinsically Disordered Regions and Proteins. Chem. Rev, 2014. 114(13): p. 6589–6631. - PMC - PubMed
-
- Honjo K, Black SE, and Verhoeff N, Alzheimer’s Disease, Cerebrovascular Disease, and the beta-amyloid Cascade. Canadian Journal of Neurological Sciences, 2012. 39(6): p. 712–728. - PubMed
-
- Walsh DM and Selkoe DJ, Oligomers in the brain: The emerging role of soluble protein aggregates in neurodegeneration. Protein Pept. Lett, 2004. 11(3): p. 213–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

