Mapping the transition state for a binding reaction between ancient intrinsically disordered proteins
- PMID: 33454008
- PMCID: PMC7762952
- DOI: 10.1074/jbc.RA120.015645
Mapping the transition state for a binding reaction between ancient intrinsically disordered proteins
Abstract
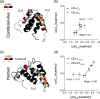
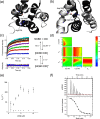
Intrinsically disordered protein domains often have multiple binding partners. It is plausible that the strength of pairing with specific partners evolves from an initial low affinity to a higher affinity. However, little is known about the molecular changes in the binding mechanism that would facilitate such a transition. We previously showed that the interaction between two intrinsically disordered domains, NCBD and CID, likely emerged in an ancestral deuterostome organism as a low-affinity interaction that subsequently evolved into a higher-affinity interaction before the radiation of modern vertebrate groups. Here we map native contacts in the transition states of the low-affinity ancestral and high-affinity human NCBD/CID interactions. We show that the coupled binding and folding mechanism is overall similar but with a higher degree of native hydrophobic contact formation in the transition state of the ancestral complex and more heterogeneous transient interactions, including electrostatic pairings, and an increased disorder for the human complex. Adaptation to new binding partners may be facilitated by this ability to exploit multiple alternative transient interactions while retaining the overall binding and folding pathway.
Keywords: IDP; coupled binding and folding; intrinsically disordered proteins; phi value analysis; pre-steady-state kinetics; protein binding; protein complex; protein evolution; protein folding; transition state.
Copyright © 2020 © 2020 Karlsson et al. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

