Structures of SARS-CoV-2 RNA-Binding Proteins and Therapeutic Targets
- PMID: 33454715
- PMCID: PMC7900486
- DOI: 10.1159/000513686
Structures of SARS-CoV-2 RNA-Binding Proteins and Therapeutic Targets
Abstract
Background: The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) epidemic has resulted in thousands of infections and deaths worldwide. Several therapies are currently undergoing clinical trials for the treatment of SARS-CoV-2 infection. However, the development of new drugs and the repositioning of existing drugs can only be achieved after the identification of potential therapeutic targets within structures, as this strategy provides the most precise solution for developing treatments for sudden epidemic infectious diseases.
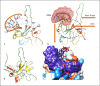
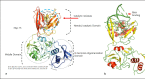
Summary: In the current investigation, crystal and cryo-electron microscopy structures encoded by the SARS-CoV-2 genome were systematically examined for the identification of potential drug targets. These structures include nonstructural proteins (Nsp-9; Nsp-12; and Nsp-15), nucleocapsid (N) proteins, and the main protease (Mpro). Key Message: The structural information reveals the presence of many potential alternative therapeutic targets, primarily involved in interaction between N protein and Nsp3, forming replication-transcription complexes (RTCs) which might be a potential drug target for effective control of current SARS-CoV-2 pandemic. RTCs consist of 16 nonstructural proteins (Nsp1-16) that play the most essential role in the synthesis of viral RNA. Targeting the physical linkage between the envelope and single-stranded positive RNA, a process facilitated by matrix proteins may provide a good alternative strategy. Our current study provides useful information for the development of new lead compounds against SARS-CoV-2 infections.
Keywords: Inhibitors; Mpro; Nsp-15; Nsp-9; Nsp12; Targets.
© 2021 S. Karger AG, Basel.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
SARS-CoV-2 nucleocapsid and Nsp3 binding: an in silico study.Arch Microbiol. 2021 Jan;203(1):59-66. doi: 10.1007/s00203-020-01998-6. Epub 2020 Aug 4. Arch Microbiol. 2021. PMID: 32749662 Free PMC article.
-
Comparison of Binding Site of Remdesivir and Its Metabolites with NSP12-NSP7-NSP8, and NSP3 of SARS CoV-2 Virus and Alternative Potential Drugs for COVID-19 Treatment.Protein J. 2020 Dec;39(6):619-630. doi: 10.1007/s10930-020-09942-9. Epub 2020 Nov 13. Protein J. 2020. PMID: 33185784 Free PMC article.
-
A Biochemical and Biophysical Analysis of the Interaction of nsp9 with nsp12 from SARS-CoV-2-Implications for Future Drug Discovery Efforts.Proteins. 2024 Nov;92(11):1308-1317. doi: 10.1002/prot.26725. Epub 2024 Jul 3. Proteins. 2024. PMID: 38958516 Free PMC article.
-
Targeting viral proteins for restraining SARS-CoV-2: focusing lens on viral proteins beyond spike for discovering new drug targets.Expert Opin Drug Discov. 2023 Mar;18(3):247-268. doi: 10.1080/17460441.2023.2175812. Epub 2023 Feb 20. Expert Opin Drug Discov. 2023. PMID: 36723288 Review.
-
Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19.Cells. 2021 Apr 6;10(4):821. doi: 10.3390/cells10040821. Cells. 2021. PMID: 33917481 Free PMC article. Review.
Cited by
-
Leukocyte metabolism in obese type 2 diabetic individuals associated with COVID-19 severity.Front Microbiol. 2022 Nov 3;13:1037469. doi: 10.3389/fmicb.2022.1037469. eCollection 2022. Front Microbiol. 2022. PMID: 36406408 Free PMC article. Review.
-
Post-COVID-19 Parkinsonism and Parkinson's Disease Pathogenesis: The Exosomal Cargo Hypothesis.Int J Mol Sci. 2022 Aug 28;23(17):9739. doi: 10.3390/ijms23179739. Int J Mol Sci. 2022. PMID: 36077138 Free PMC article. Review.
-
Genetic Engineering Systems to Study Human Viral Pathogens from the Coronaviridae Family.Mol Biol. 2022;56(1):72-89. doi: 10.1134/S0026893322010022. Epub 2022 Feb 12. Mol Biol. 2022. PMID: 35194246 Free PMC article.
-
Molecular characteristics, immune evasion, and impact of SARS-CoV-2 variants.Signal Transduct Target Ther. 2022 Jun 28;7(1):202. doi: 10.1038/s41392-022-01039-2. Signal Transduct Target Ther. 2022. PMID: 35764603 Free PMC article. Review.
-
Potential Coronaviral Inhibitors of the Nucleocapsid Protein Identified In Silico and In Vitro from a Large Natural Product Library.Pharmaceuticals (Basel). 2022 Aug 24;15(9):1046. doi: 10.3390/ph15091046. Pharmaceuticals (Basel). 2022. PMID: 36145267 Free PMC article.
References
-
- Nur SM, Hasan MA, Amin MA, Hossain M, Sharmin T. Design of potential RNAi (miRNA and siRNA) molecules for Middle East respiratory syndrome coronavirus (MERS-CoV) gene silencing by computational method. Interdiscip Sci. 2014 Nov - PubMed
-
- Khan U, Mehta R, Arif M, Lakhani O. Pandemics of the past: a narrative review. J Pak Med Assoc. 2020;((0)):1. - PubMed
-
- Greene WC. A history of AIDS: looking back to see ahead. Eur J Immunol. 2007 Nov;37((Suppl 1)):S94–102. - PubMed
-
- Fauci AS, Marston HD. Ending the HIV-AIDS pandemic: follow the science. N Engl J Med. 2015 Dec;373((23)):2197–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

