The genetic landscape of polycystic kidney disease in Ireland
- PMID: 33454723
- PMCID: PMC8110806
- DOI: 10.1038/s41431-020-00806-5
The genetic landscape of polycystic kidney disease in Ireland
Abstract
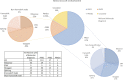
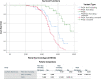
Polycystic kidney diseases (PKDs) comprise the most common Mendelian forms of renal disease. It is characterised by the development of fluid-filled renal cysts, causing progressive loss of kidney function, culminating in the need for renal replacement therapy or kidney transplant. Ireland represents a valuable region for the genetic study of PKD, as family sizes are traditionally large and the population relatively homogenous. Studying a cohort of 169 patients, we describe the genetic landscape of PKD in Ireland for the first time, compare the clinical features of patients with and without a molecular diagnosis and correlate disease severity with autosomal dominant pathogenic variant type. Using a combination of molecular genetic tools, including targeted next-generation sequencing, we report diagnostic rates of 71-83% in Irish PKD patients, depending on which variant classification guidelines are used (ACMG or Mayo clinic respectively). We have catalogued a spectrum of Irish autosomal dominant PKD pathogenic variants including 36 novel variants. We illustrate how apparently unrelated individuals carrying the same autosomal dominant pathogenic variant are highly likely to have inherited that variant from a common ancestor. We highlight issues surrounding the implementation of the ACMG guidelines for variant pathogenicity interpretation in PKD, which have important implications for clinical genetics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- ERA-EDTA Registry Committee. ERA-EDTA Registry Annual Report 2017. 2017.
-
- Audrézet MP, Cornec-Le Gall E, Chen JM, Redon S, Quéré I, Creff J, et al. Autosomal dominant polycystic kidney disease: comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Hum Mutat. 2012;33:1239–50. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

