Antimicrobial resistance clusters in commensal Escherichia coli from livestock
- PMID: 33455079
- PMCID: PMC8048968
- DOI: 10.1111/zph.12805
Antimicrobial resistance clusters in commensal Escherichia coli from livestock
Abstract
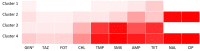
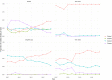
To combat antimicrobial resistance (AMR), policymakers need an overview of evolution and trends of AMR in relevant animal reservoirs, and livestock is monitored by susceptibility testing of sentinel organisms such as commensal E. coli. Such monitoring data are often vast and complex and generates a need for outcome indicators that summarize AMR for multiple antimicrobial classes. Model-based clustering is a data-driven approach that can help to objectively summarize AMR in animal reservoirs. In this study, a model-based cluster analysis was carried out on a dataset of minimum inhibitory concentrations (MIC), recoded to binary variables, for 10 antimicrobials of commensal E. coli isolates (N = 12,986) derived from four animal species (broilers, pigs, veal calves and dairy cows) in Dutch AMR monitoring, 2007-2018. This analysis revealed four clusters in commensal E. coli in livestock containing 201 unique resistance combinations. The prevalence of these combinations and clusters differs between animal species. Our results indicate that to monitor different animal populations, more than one indicator for multidrug resistance seems necessary. We show how these clusters summarize multidrug resistance and have potential as monitoring outcome indicators to benchmark and prioritize AMR problems in livestock.
Keywords: E. coli; antimicrobial drug resistance; cluster analysis; epidemiological monitoring; multidrug resistance; policy making.
© 2021 The Authors. Zoonoses and Public Health published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Biernacki, C. , Celeux, G. , & Govaert, G. (2000). Assessing a mixture model for clustering with the integrated completed likelihood. IEEE Transactions on Pattern Analysis and Machine Intelligence, 22(7), 719–725. 10.1109/34.865189 - DOI
-
- Bos, M. E. H. , Mevius, D. J. , Wagenaar, J. A. , van Geijlswijk, I. M. , Mouton, J. W. , & Heederik, D. J. J. & Netherlands Veterinary Medicines Authority (SDa) (2015). Antimicrobial prescription patterns of veterinarians: Introduction of a benchmarking approach. Journal of Antimicrobial Chemotherapy, 70(8), 2423–2425. 10.1093/jac/dkv104 - DOI - PubMed
-
- Buyle, F. , Metz‐Gercek, S. , Mechtler, R. , Kern, W. , Robays, H. , Vogelaers, D. , & Struelens, M. (2013). Development and validation of potential structure indicators for evaluating antimicrobial stewardship programs in European hospitals. European Journal of Clinical Microbiology & Infectious Diseases., 32, 1161–1170. 10.1007/s10096-013-1862-4 - DOI - PubMed
-
- Chantziaras, I. , Smet, A. , Filippitzi, M. E. , Damiaans, B. , Haesebrouck, F. , Boyen, F. , & Dewulf, J. (2018). The effect of a commercial competitive exclusion product on the selection of enrofloxacin resistance in commensal E. coli in broilers. Avian Pathology, 47(5), 443–454. 10.1080/03079457.2018.1486027 - DOI - PubMed
-
- Dorado‐Garcia, A. , Mevius, D. J. , Jacobs, J. J. , Van Geijlswijk, I. M. , Mouton, J. W. , Wagenaar, J. A. , & Heederik, D. J. (2016). Quantitative assessment of antimicrobial resistance in livestock during the course of a nationwide antimicrobial use reduction in the Netherlands. Journal of Antimicrobial Chemotherapy, 71(12), 3607–3619. 10.1093/jac/dkw308 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

