STAT3/miR-15a-5p/CX3CL1 Loop Regulates Proliferation and Migration of Vascular Endothelial Cells in Atherosclerosis
- PMID: 33456354
- PMCID: PMC7807201
- DOI: 10.7150/ijms.49460
STAT3/miR-15a-5p/CX3CL1 Loop Regulates Proliferation and Migration of Vascular Endothelial Cells in Atherosclerosis
Abstract
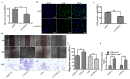
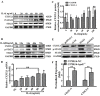
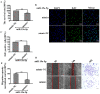
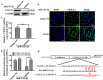
Endothelial cell proliferation disorder caused by vascular injury seems to be one of the causes of atherosclerosis, which is the pathological basis of coronary heart disease. The role of STAT3 in the regulation of microRNAs and endothelial dysfunction in atherosclerosis is unclear. STAT3 can be activated by cytokine IL-6 and up regulate the expression of CX3CL1. In addition, microRNA-15a-5p (miR-15a-5p) inhibited the transcription of CX3CL1, the proliferation of vascular endothelial cells and the proliferation of STAT3 regulated vascular endothelial cells. STAT3 positively regulates the expression of CX3CL1, and then down-regulates the inhibition of CX3CL1 by over-expression of miR-15a-5p, thus forming an elimination feedback loop to control the proliferation of HUVECs and affect the progression of atherosclerosis. In conclusion, miR-15a-5p may be the therapeutic target of the pathological basis of coronary atherosclerosis.
Keywords: Atherosclerosis; CX3CL1; Endothelial cells; MicroRNA-15a; STAT3.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
References
-
- Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–809. - PubMed
-
- Clark AK, Staniland AA, Marzia M. Fractalkine/CX3CR1 signalling in chronic pain and inflammation. Current Pharmaceutical Biotechnology. 2011;12:1707–14. - PubMed
-
- Edsfeldt A, Grufman H, Asciutto G. et al. Circulating cytokines reflect the expression of pro-inflammatory cytokines in atherosclerotic plaques. Atherosclerosis. 2015;241:443–9. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

