SMRT-based mitochondrial genome of the edible mushroom Morchella conica
- PMID: 33458112
- PMCID: PMC7782166
- DOI: 10.1080/23802359.2020.1810160
SMRT-based mitochondrial genome of the edible mushroom Morchella conica
Abstract
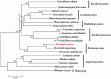
Morchella conica Pers. is a highly-prized mushroom for its edible and medical values. In this study, we determined the complete mitochondrial genome of M. conica combining both PacBio and Illumina sequencing technologies. The complete mitochondrial genome is 280,763 bp in length with a GC content of 39.88%. We identified a total of 14 core conserved protein-coding genes, 127 non-conserved open reading frames (ncORFs) and 30 tRNA genes in the M. conica mitogenome. However, no large or small rRNA subunits (rnl or rns) were identified in this mitogenome. In addition, we detected two mitochondrial RNase P (rnpB) genes and one ribosomal protein genes (rps3). Phylogenetic analysis was performed among M. conica and 18 other representative fungi from Ascomycota, Basidiomycota and Mucoromycota. Our results showed that M. conica was most closely related to M. importuna. The availability of the M. conica mitochondrial genome will form the basis of genetic breeding program and enhance our understanding of the evolution of this species.
Keywords: Morchella conica; PacBio; evolution; mitochondrial genome; mushroom.
© 2020 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
The mitochondrial genome of Morchella importuna (272.2 kb) is the largest among fungi and contains numerous introns, mitochondrial non-conserved open reading frames and repetitive sequences.Int J Biol Macromol. 2020 Jan 15;143:373-381. doi: 10.1016/j.ijbiomac.2019.12.056. Epub 2019 Dec 9. Int J Biol Macromol. 2020. PMID: 31830457
-
Subchromosome-Scale Nuclear and Complete Mitochondrial Genome Characteristics of Morchella crassipes.Int J Mol Sci. 2020 Jan 12;21(2):483. doi: 10.3390/ijms21020483. Int J Mol Sci. 2020. PMID: 31940908 Free PMC article.
-
The complete mitochondrial genome of the Basidiomycete fungus Pleurotus cornucopiae (Paulet) Rolland.Mitochondrial DNA B Resour. 2018 Jan 3;3(1):73-75. doi: 10.1080/23802359.2017.1422405. Mitochondrial DNA B Resour. 2018. PMID: 33490488 Free PMC article.
-
A Polysaccharide Purified from Morchella conica Pers. Prevents Oxidative Stress Induced by H₂O₂ in Human Embryonic Kidney (HEK) 293T Cells.Int J Mol Sci. 2018 Dec 13;19(12):4027. doi: 10.3390/ijms19124027. Int J Mol Sci. 2018. PMID: 30551572 Free PMC article.
-
A comprehensive review on Morchella importuna: cultivation aspects, phytochemistry, and other significant applications.Folia Microbiol (Praha). 2021 Apr;66(2):147-157. doi: 10.1007/s12223-020-00849-7. Epub 2021 Jan 19. Folia Microbiol (Praha). 2021. PMID: 33464471 Review.
Cited by
-
Transcriptomic and Metabolomic Profiles Provide Insights into the Red-Stipe Symptom of Morel Fruiting Bodies.J Fungi (Basel). 2023 Mar 18;9(3):373. doi: 10.3390/jof9030373. J Fungi (Basel). 2023. PMID: 36983541 Free PMC article.
-
Mating Systems in True Morels (Morchella).Microbiol Mol Biol Rev. 2021 Aug 18;85(3):e0022020. doi: 10.1128/MMBR.00220-20. Epub 2021 Jul 28. Microbiol Mol Biol Rev. 2021. PMID: 34319143 Free PMC article. Review.
-
Insights into Fungal Mitochondrial Genomes and Inheritance Based on Current Findings from Yeast-like Fungi.J Fungi (Basel). 2024 Jun 21;10(7):441. doi: 10.3390/jof10070441. J Fungi (Basel). 2024. PMID: 39057326 Free PMC article. Review.
-
Characterization of the mitochondrial genomes of three powdery mildew pathogens reveals remarkable variation in size and nucleotide composition.Microb Genom. 2021 Dec;7(12):000720. doi: 10.1099/mgen.0.000720. Microb Genom. 2021. PMID: 34890311 Free PMC article.
References
-
- Elmastas M, Isildak O, Turkekul I, Temur N.. 2007. Determination of antioxidant activity and antioxidant compounds in wild edible mushrooms. J Food Compost Anal. 20(3–4):337–345.
-
- Gençcelep H, Uzun Y, Tunçtürk Y, Demirel K.. 2009. Determination of mineral contents of wild-grown edible mushrooms. Food Chem. 113(4):1033–1036.
-
- Kalač P. 2009. Chemical composition and nutritional value of European species of wild growing mushrooms: a review. Food Chem. 113(1):9–16.
LinkOut - more resources
Full Text Sources
Miscellaneous
